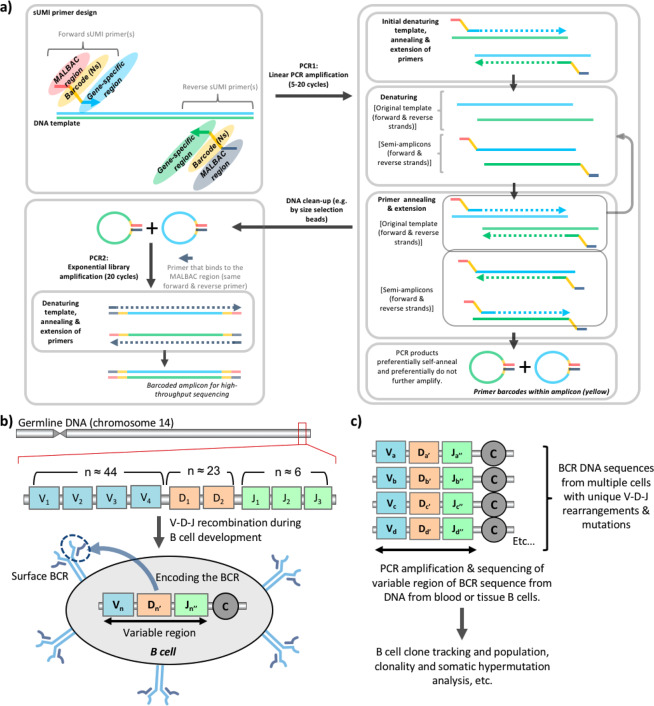
Figure 1.
Schematic of the sUMI-seq PCR with DNA molecular barcoding approach. (a) Schematic diagram of sUMI-seq. Quantification of pools of DNA variants can be important across multiple fields in biology, including B cell receptor (BCR) repertoire sequencing. MALBAC-barcoded primer design is shown. Amplicons from PCR1, after a clean-up step, are amplified in PCR2 using forward primers priming gene specific regions (as in PCR1) and reverse universal primers binding to the MALBAC regions (b) Schematic diagram of BCR rearrangement: B cells are generated from haematopoietic stem cells. The IGH gene locus on chromosome 14 (in humans) encodes for multiple distinct copies of the variable (V), diversity (D), and joining (J) genes, with functional IGH BCR (one functional allele per cell) generated during B cell differentiation by site-specific V-D-J recombination. Random deletions and insertions of nucleotides during recombination results in sequence diversification at the gene junctional regions. Rearranged BCR genes can be further diversified through somatic hypermutation (SHM) upon B cell activation. (c) A BCR repertoire is defined as the BCRs collection from a B cell population, such as from blood or tissues. B cell DNA can be used to construct BCR libraries, using multiplex PCR with forward primers annealing to all the variable VH gene families (IgH V1–7) and reverse primers annealing to the JH gene families (IgH J1-6).