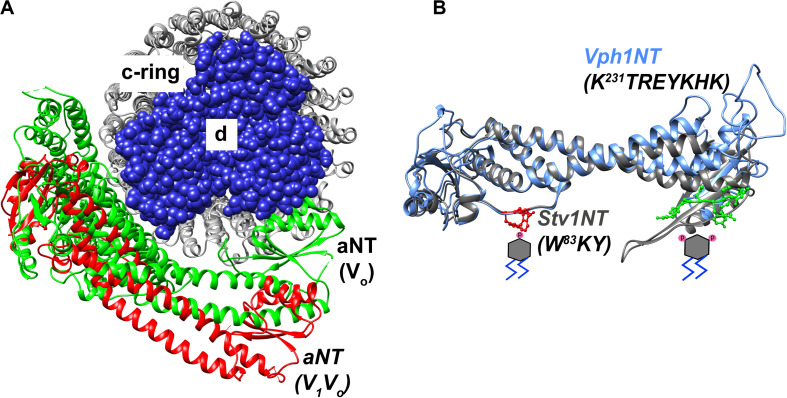
FIGURE 4.
Structural features of the a-subunit. (A) A cryo-EM based model indicating the dynamic structural re-orientation of the N-terminal domain of the a-subunit in an assembled and active holoenzyme-conformation, colored in red (Zhao et al., 2015), and a disassembled and autoinhibited conformation in the isolated Vo sector, colored in green (Stam and Wilkens, 2017). The chain traces of the c-ring comprized of c8c′c″ is colored in gray and the spherical atoms of the d-subunit is colored in blue. (B) A superimposition of the cryo-EM structure of the NT-domain of Vph1 in blue (Zhao et al., 2015) and a Phyre2 based structural model of the Stv1-NT in gray. Stv1NT was modeled to the available low-density structure of the Stv1-V-ATPase (Vasanthakumar et al., 2019). PI(4)P and PI(3,5)P2 molecules are indicated with phosphates drawn in pink on the respective positions of a phosphatidylinositol lipid below the proximal and distal subdomains of the aNT domain, respectively. The PI(4)P binding site in Stv1, comprized of a W83KY sequence in the proximal end of the Stv1NT domain is indicated using red ball-and-stick atomic side chains. A PI(3,5)P2 recognition site in Vph1, comprized of a K231TREYKHK sequence in the distal end of the Vph1NT domain is indicated using green ball-and-stick atomic side chains.