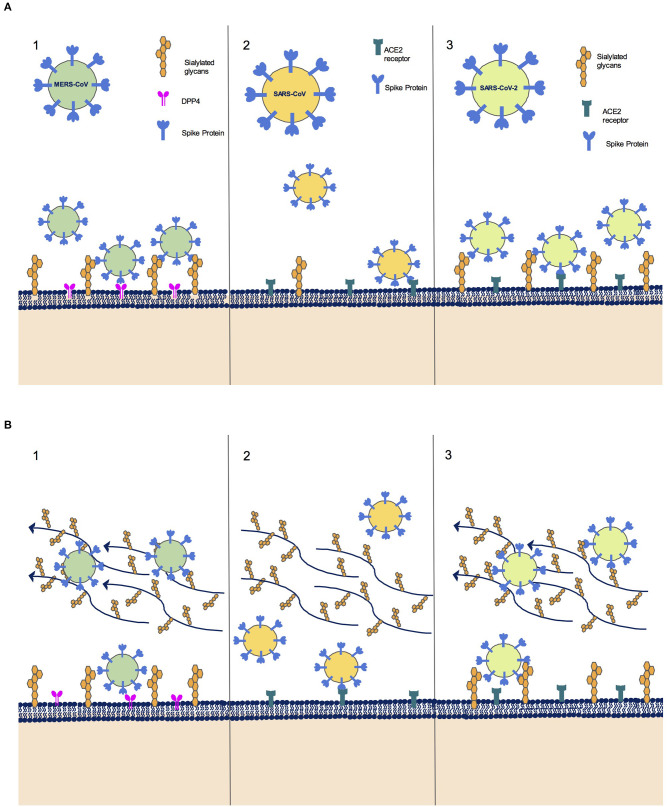
Figure 1.
(A) Sialic acid recognition as an infection facilitator for Coronavirus strains. (1) MERS-CoV binds to non-acetylated sialoside receptors on the epithelial cells of the respiratory tract, promoting clustering and facilitating its binding to its receptor DPP4. (2) SARS-CoV binds to ACE2 receptor. (3) SARS-CoV-2 binds to ACE2 receptor, but a surface region in Spike protein is very similar to MERS-CoV spike sialic acid-binding region, suggesting a possible role of sialic acid recognition in infection initiation. (B) Sialic acid recognition as a host defense mechanism for Coronavirus strains. (1) MERS-CoV can bind to sialylated O-linked glycans covering mucins on mucosal cell surfaces, thus being trapped in the mucous layer and consequently eliminated through ciliary movement. (2) SARS-CoV passes through the mucous layer without being stopped by decoy alternative binding sites. (3) SARS-CoV-2 shares with MERS-CoV the sialic acid binding region of Spike protein, and could therefore bind to sialylated O-linked glycans similarly to MERS-CoV, thus possibly being eliminated through ciliary movement.