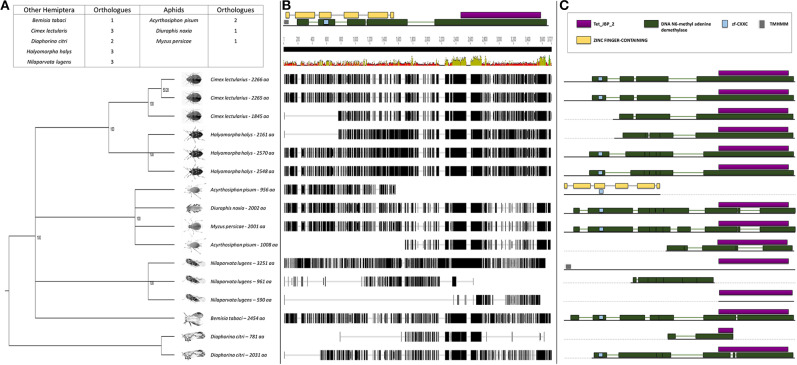
Figure 4.
(A) Phylogenetic analysis of the N6-methyl adenine demethylase (TET-like) amino acid sequences from eight Hemipterans species using MAFFT v7.4 and Paup v4.0a136. Included in the analysis are Acyrthosiphon pisum, Bemisia tabaci, Cimex lectularius, Diaphorina citri, Diuraphis noxia, Halyomorpha halys, Myzus persicae, and Nilaparvata lugens; (B) Comparison of the N6-methyl adenine demethylase (TET-like) gene sequence from the different hemipteran species in (A) showing the level of conservation on gene sequence level; and (C) Comparison of the conservancy on functional motifs within the N6-methyl adenine demethylase (TET-like) gene sequence from the different hemipteran species in (A). Indicated are Tet_JBP_2, DNA N6-methyl adenine demethylase, zf-CXXC, TMHMM, and Zinc finger-containing motifs.