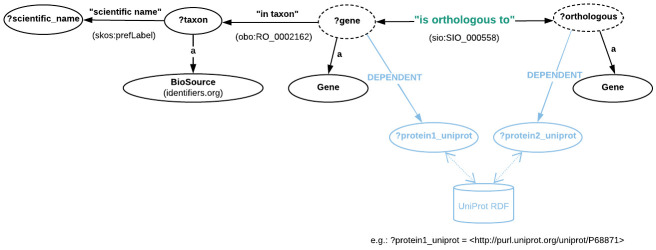
Figure 5. Directed graph abstraction of a portion of the EBI RDF graph related to pairwise orthologous genes.
Moreover, as opposed to the RDF representations in OMA and MBGD, here the pairwise orthology is explicitly asserted through the “is orthologous to” property (more precisely, http://semanticscience.org/resource/SIO_000558) as shown in Figure 5. However, there is no information available regarding orthologous clusters. Moreover, the Gene class here is in fact the OBO (not ORTH) class, i.e. http://purl.obolibrary.org/obo/SO_0000704. Instances of these genes can be specified either through their cross-reference to UniProt (the http://rdf.ebi.ac.uk/terms/ensembl/DEPENDENT property) or directly through their ENSEMBL identifier, by fixing the value of ?gene to the concatenation of http://rdf.ebi.ac.uk/resource/ensembl/ and the corresponding Ensembl identifier. Finally, the taxonomic identifiers are provided via instances of the BioSource class, http://www.biopax.org/release/biopax-level3.owl#BioSource.