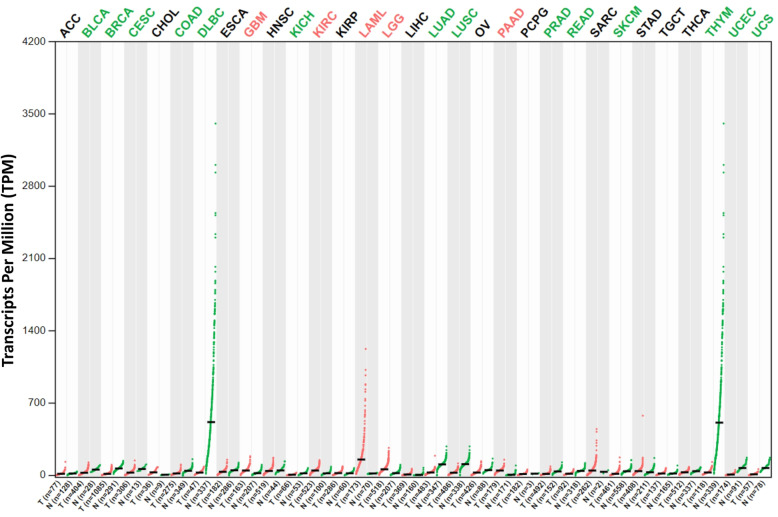
Fig. 3.
Expression profile analyses of VISTA across multiple cancers and normal tissues. Expression pattern of VISTA in ACC, BLCA, BRCA, CESC, CHOL, COAD, DLBC, ESCA, GBM, HNSC, KICH, KIRC, KIRP, LAML, LGG, LIHC, LUAD, LUSC, OV, PAAD, PCPG, PRAD, READ, SARC, SKCM, STAD, TGCT, THCA, THYM, UCEC, and UCS. GEPIA was used to generate dot plots profiling VISTA expression patterns across multiple cancer types (TCGA tumor) and paired normal tissue samples (TCGA normal + GTEx normal). Each dot represents the individual expression of a distinct tumor or normal sample. ANOVA method was used for differential gene expression analysis, and genes with higher |log2FC| values (> 1) and lower q values (< 0.01) were considered differentially expressed genes