Figure 5. NMDA receptor structure and ligand binding sites.
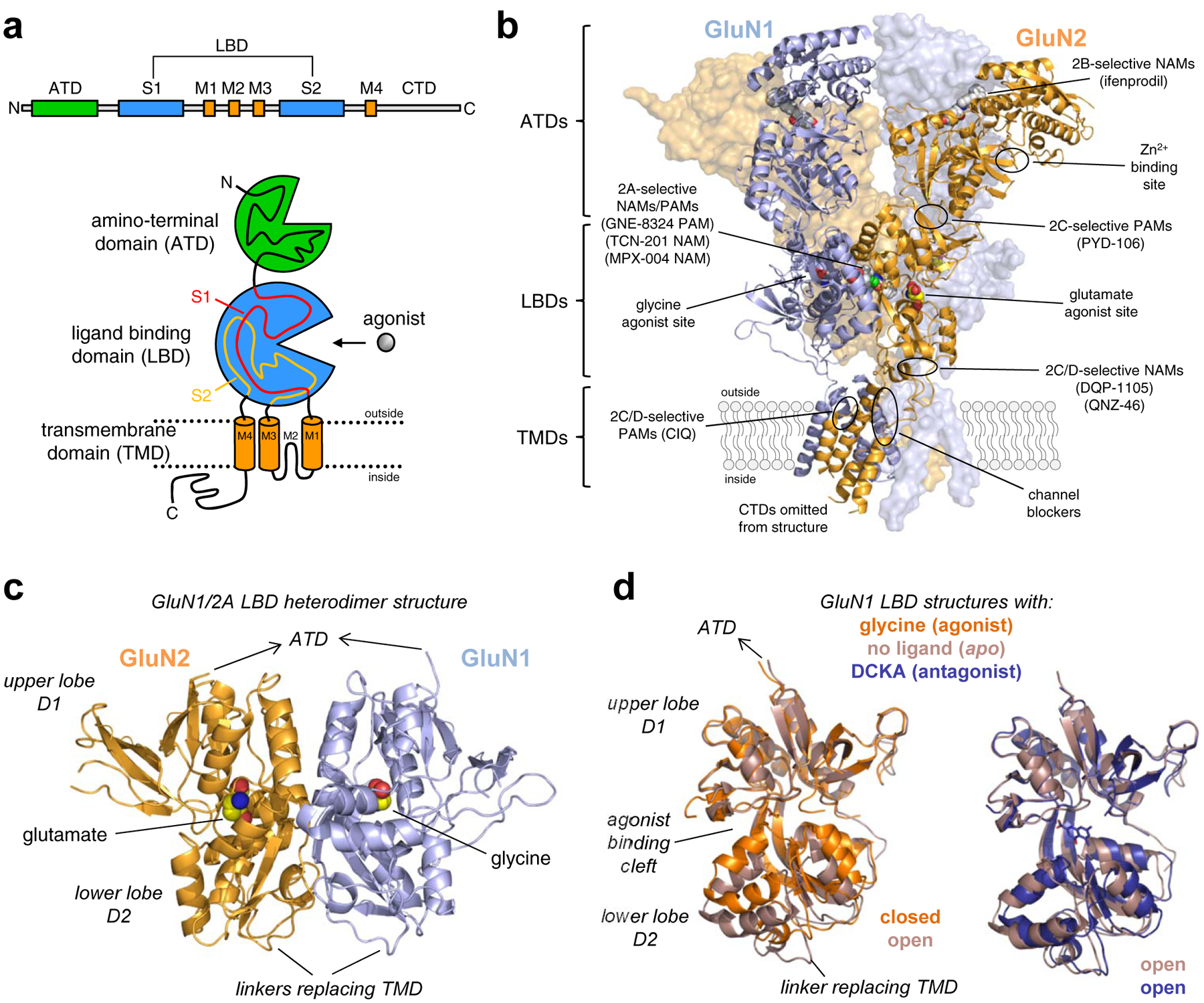
a) Linear representation and cartoon illustration of the polypeptide chain in iGluR subunits. Each subunit consists of a large extracellular amino-terminal domain (ATD), a bi-lobed ligand binding domain (LBD), a transmembrane domain (TMD), and an intracellular CTD. The TMD is formed by three transmembrane helices (M1, M2, and M4) and a membrane re-entrant loop (M2). The LBD is formed by two segments of the polypeptide chain (S1 and S2), which fold into a kidney-shaped structure composed of an upper lobe (D1) and lower lobe (D2) relative to the cell membrane, and the agonist binding site is located in the cleft between the two lobes. b) Crystal structure of the GluN1/2B NMDA receptor (PDB ID 4PE5; [67]), illustrating the subunit arrangement and the layered domain organization composed of the TMD layer and two extracellular layers formed by LBDs and ATDs. Agonist binding sites as well as known and predicted binding sites for positive and negative allosteric modulators (PAMs and NAMs) are highlighted. c) Crystal structure of the soluble GluN1/2A LBD heterodimer (PDB ID 5I57; [158]), showing the subunit interface and back-to-back dimer arrangement of the LBDs. Soluble LBD proteins composed of the S1 and S2 segments of the polypeptide chain are produced by deleting the ATD and replacing the TMD with a di-peptide linker. d) Overlay of crystal structures of the soluble GluN1 LBD in the apo-form (PDB 4KCC; [168]) or in complex with the agonist glycine (PDB ID 5I57; [158]) or competitive antagonist DCKA (PDB ID 4NF4; [166]). The upper D1 lobes are aligned to illustrate the similar conformations of antagonist-bound and apo-form structures. Agonist binding induces considerable closure of the LBD compared to the antagonist-bound and apo-form structures, and agonist-induced closure of the LBD is required for activation of NMDA receptors. Competitive antagonists bind the LBD without inducing domain closure, thereby preventing agonist binding and receptor activation.
