Figure 3: HSV-1 miR-H16 targets FIH-1 to activate NOTCH signaling.
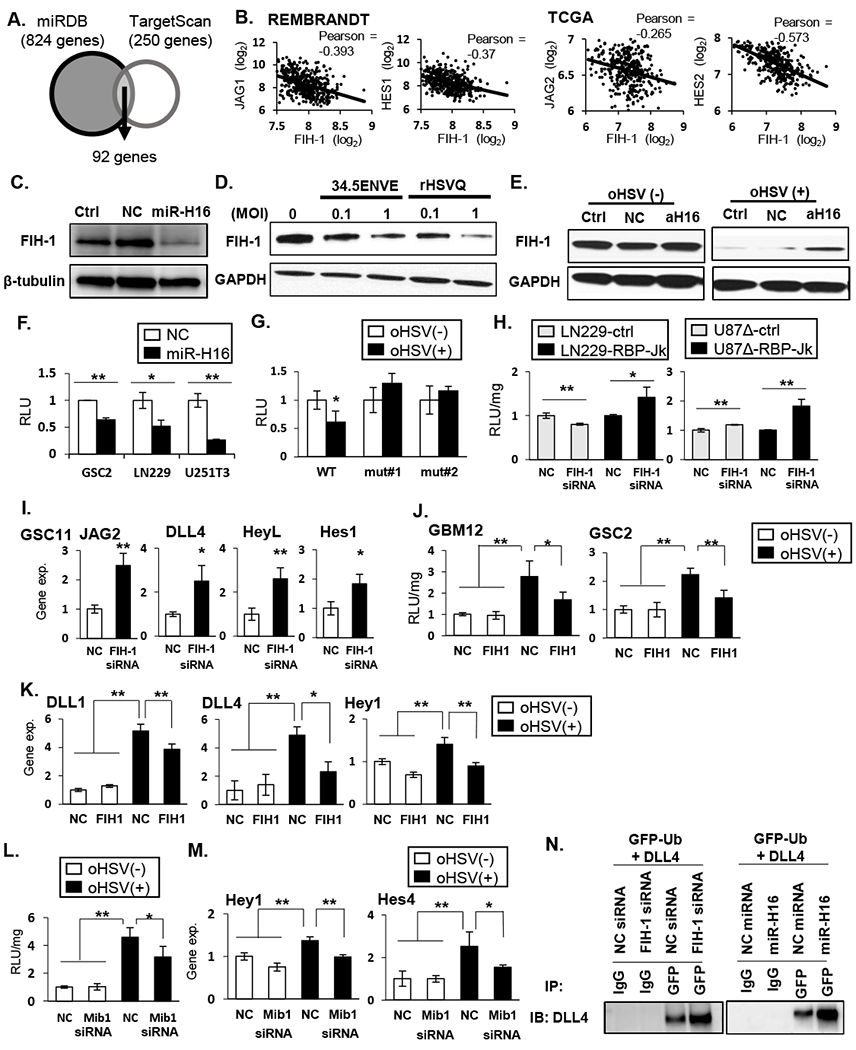
A: Schematic of overlap between genes predicted to be regulated by microRNA-H16 (miR-H16) in miRDB and TargetScan. B: Scatterplot of two-gene correlation between FIH-1 and NOTCH ligands and downstream target genes in REMBRANDT and TCGA. C: Western blot for FIH-1 expression in LN229 cells transfected with non-targeting control miRNA (NC) or miR-H16 (H16). D: Western blot analysis evaluating changes in FIH-1 after infection with 34.5 ENVE or rHSVQ. E: Antagomir for miR-H16 (amiR-H16) was transfected in LN229 cells 48 hours prior to infection with 34.5ENVE and probed for FIH-1 expression. F-G: FIH-1 3’UTR assay. Luciferase upstream of 3’ UTR sequence of FIH-1 containing the predicted wild type (WT) or mutant (mut#1 and mut#2) miR-H16 target site were transfected in indicated glioma cells. Relative changes in mean luciferase activity after miR-H16 transfection (F) or 34.5ENVE infection (G) is shown (n ≥ 3). H-I: knockdown of FIH-1 induced NOTCH reporter activity (H) and NOTCH target gene expression (I). J-K: Overexpression of FIH-1 reduced oHSV-induced NOTCH activity (J) and NOTCH target gene expression (K) (n≥ 3). L-M: Knockdown of mind bomb 1 (Mib1) by siRNA reduced virus induced NOTCH reporter activity (L) and target gene expression (M) in GBM12. Data are the mean ± SD of the relative change (n=4). N: Ubiquitylation of DLL4 in the presence of FIH-1 siRNA (left panel) or miR-H16 (right panel). Cells were transfected with GFP-tagged Ubiquitin. FIH-1 siRNA and miR-H16 increased DLL4 interaction with ubiquitin. *P<0.05, **P<0.01.
