FIGURE 2.
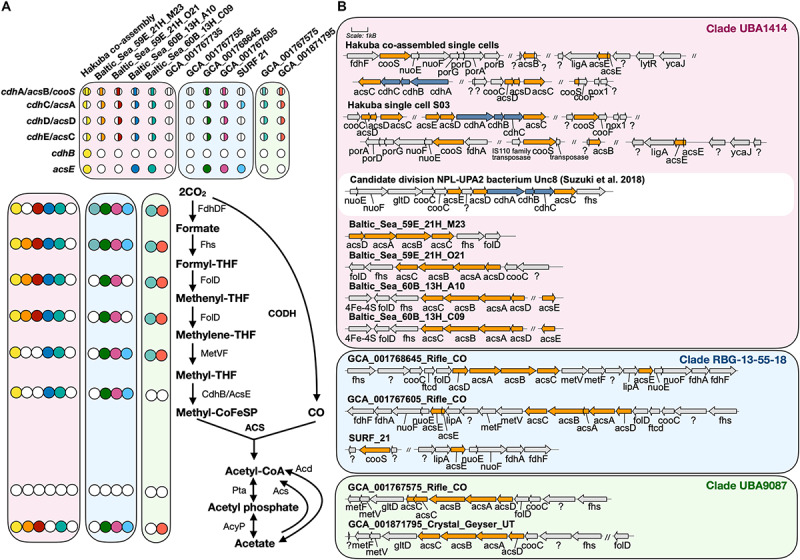
Key enzymes encoded by genomes within clades UBA1414, RBG-13-55-18, and UBA9087 for the Wood–Ljungdahl Pathway. (A) The presence of proteins involved in the WL pathway are depicted with a colored circle while the absence is depicted by a white circle. Each color corresponds to a genome used in this study. For CODH/ACS, the colored circle is split by the bacterial- or archaeal-type subunit, and the location of the color indicates whether a gene was present or not within the assembly. The proteins Acs and Acd do not have corresponding circles. (B) The gene neighborhood of CODH/ACS is shown for each genome and was designed using Gene Graphics (Harrison et al., 2017). Gene neighborhoods on different contigs are denoted by “//.” The three clades are color coded as done in Figure 1. The gene neighborhood of Candidate division NPL-UPA2 bacterium Unc8 (Suzuki et al., 2018), a putative acetogen from a serpentinite-hosted system called The Cedars, is depicted in a white box for comparison of hybrid CODH/ACS. Abbreviations: acyP (acylphosphatase), acd (ADP-forming acetyl-CoA synthetase), acs (acetyl-CoA synthase), cdh (carbon monoxide dehydrogenase), cooS (carbon monoxide dehydrogenase), fdh (formate dehydrogenase), fhs [formyl-tetrahydrofolate (THF) synthase], folD (bifunctional 5,10-methenyl-THF cyclohydrolase/5,10-methylene-THF dehydrogenase), ftcD (glutamate formiminotransferase/formiminotetrahydrofolate cyclodeaminase), gltD (glutamate synthase), ligA (DNA ligase), lipA (lipoyl synthase), lytR (cell envelope-related function), met (Methylene-THF reductase), nox1 (NADH oxidase), nuo (NADH-quinone oxidoreductase), pta (phosphotransacetylase), por (pyruvate ferredoxin oxidoreductase), ycaJ (putative ATPase).
