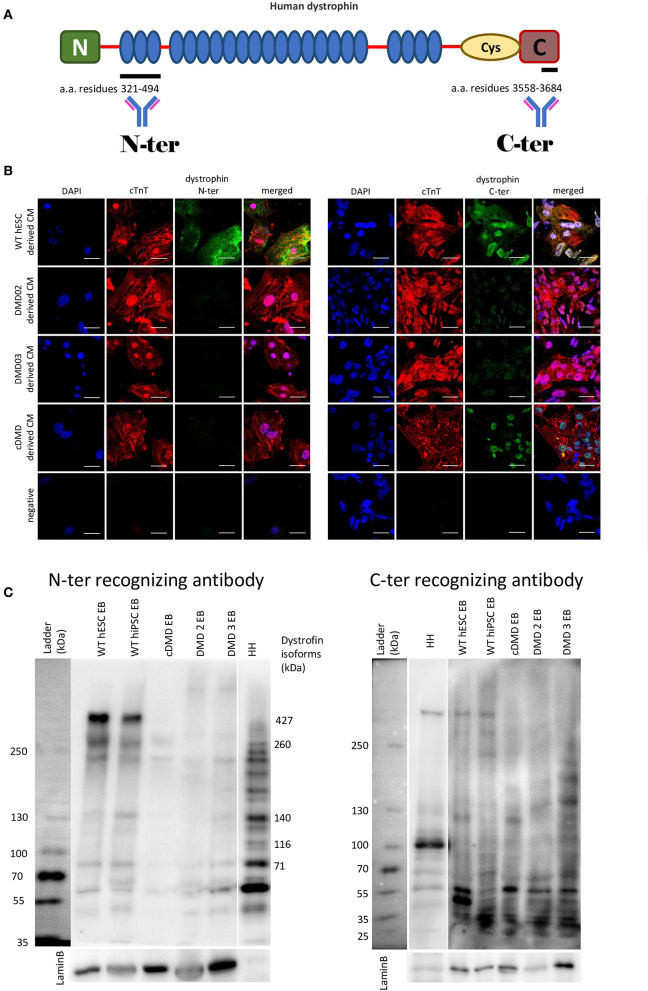
Figure 2.
DMD-hPSC differentiate into cardiac cells (CCs), which do not express dystrophin. (A) Antibodies against dystrophin N-terminal (N) aminoacid (a.a.) residues 321–494 and C-terminal aminoacid residues 3558–3684 were used. (B) hPSC derived CCs were analyzed for presence of dystrophin (in green). WT hESC derived CCs dissociated from spontaneously beating EBs presenting with positive signal for cardiac troponin T (red) showed positive signal for dystrophin labeled by N-terminus specific antibody as well as by C-terminus specific antibody. None of the DMD-hPSC lines (DMD02, DMD03, and cDMD) derived CCs showed any signal for dystrophin. DAPI was used for nuclear counterstain. Ruler represents 50 μm. (C) The differentiated EBs were lyzed and western blot analysis was performed to identify the individual isoforms expressed. Both used WT-hPSC derived EBs show expression of various dystrophin isoforms including Dp427, Dp260, Dp140, and Dp71. The CRISPR/Cas9 generated cDMD-EBs show no expression of either isoform, while only shorter isoforms were identified in patient DMD-hiPSC-EBs with promoters downstream of the patient's mutations. Human heart tissue (HH) was used as positive control. LaminB was used as loading control.