FIGURE 5.
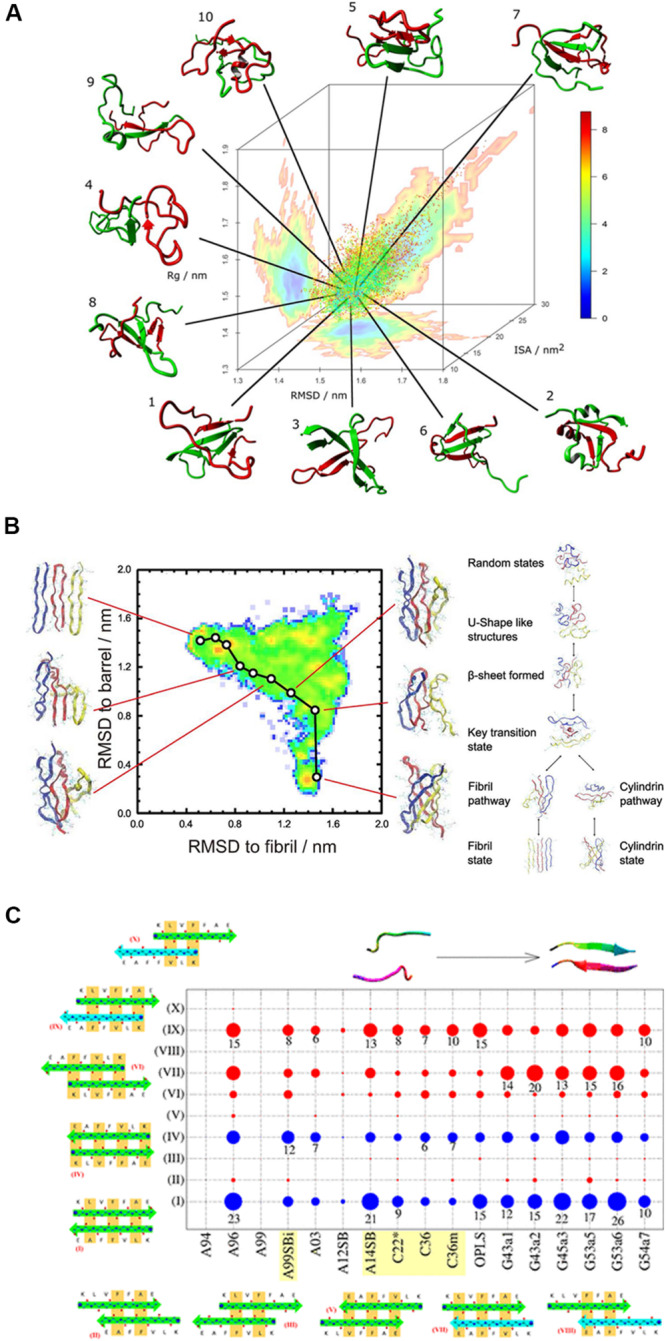
(A) A typical REMD simulation that shows the dimerization of the Aβ1–40 peptide and the free energy landscape that is projected three-dimensionally: radius of gyration (Rg), RMSD from the global average conformation and interaction surface area (ISA). (B) Understanding the transition between fibril-like and barrel-like, αB-crystalline segment using the Replica-Exchange-with-Tunneling method. A typical pathway between the two states is represented on the free energy landscape that is projected on the root-mean-square-deviation spaces. The inter-conversion mechanism among the coil structure, fibril state, and cylindrical state is proposed based on the energy landscape. (C) Comparison of 17 force fields with the Aβ16–22 dimer. The conformational populations of the Aβ16–22 dimer are shown in circles for all the tested force fields. The blue circles represent the in-register antiparallel β-sheets (I and IV) and the red ones indicate the other 8 out-of-register patterns, indicating that the H-bond patterns are shifted and are low in number. The circle size represents the population. AMBER99-ILDN, AMBER14SB, CHARMM22*, CHARMM36, and CHARMM36m are the best candidates for studying amyloid peptides.
