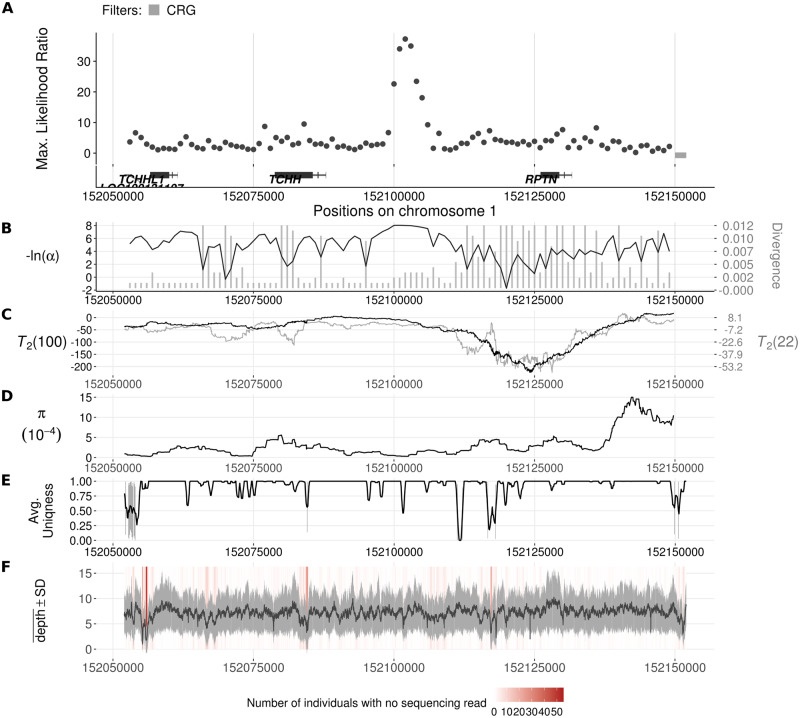
Fig 10. Introgression sweep signals, parameter estimates, and sequencing properties across the 100 kb region on chromosome 1 covering TCHH and RPTN genes in YRI.
A. Likelihood ratio test statistic computed from Model 1 of VolcanoFinder on data on within-YRI polymorphism and substitutions with respect to chimpanzee. Horizontal dark gray bars correspond to regions that were filtered based on mean CRG score. Gene tracts and labels for key genes are depicted below the plot, with the wider bars representing exons. B. Values for α and divergence D corresponding to the maximum likelihood estimate of the data. Black line corresponds to −ln(α) and vertical gray bars correspond to estimated D. C. Likelihood ratio test statistic computed from T2 of BALLET on data on within-YRI polymorphism and substitutions with respect to chimpanzee using windows of 100 (black) or 22 (gray) informative sites on either side of the test site. D. Mean pairwise sequence difference () computed in five kb windows centered on each polymorphic site. E. Mappability uniqueness scores for 35 nucleotide sequences across the region. F. Mean sequencing depth across the 108 YRI individuals as a function of genomic position, with the gray ribbon indicating standard deviation. The background heatmap displays the number of individuals devoid of sequencing reads as a function of genomic position, with darker shades of red indicating a greater number of individuals with no sequencing reads.