Abstract
Since the end of 2019, the global COVID-19 outbreak has once again made coronaviruses a hot topic. Vaccines are hoped to be an effective way to stop the spread of the virus. However, there are no clinically approved vaccines available for coronavirus infections. Reverse genetics technology can realize the operation of RNA virus genomes at the DNA level and provide new ideas and strategies for the development of new vaccines. In this review, we systematically describe the role of reverse genetics technology in studying the effects of coronavirus proteins on viral virulence and innate immunity, cell and tissue tropism and antiviral drug screening. An efficient reverse genetics platform is useful for obtaining the ideal attenuated strain to prepare an attenuated live vaccine.
Keywords: Coronavirus, Reverse genetics, Live attenuated vaccine
1. Introduction
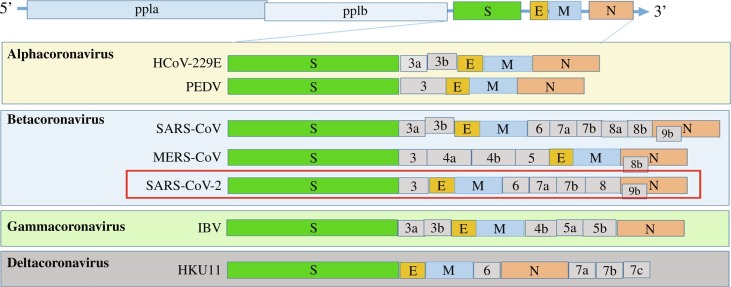
Coronaviruses are large, enveloped, positive-sense, single-stranded RNA viruses with genome sizes ranging from 26 to 32 kb that are distributed broadly among humans, other mammals, and birds and cause respiratory, enteric, hepatic, and neurologic diseases (Coronaviridae Study Group of the International Committee on Taxonomy of Viruses, 2020; Cui et al., 2019). Coronaviridae belongs to the order Nidovirales along with three other families (Arteriviridae, Mesoniviridae, and Roniviridae). Coronaviridae is further classified into four genera (Fig. 1A) based on phylogenetic analyses and genomic structures, namely, Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus (Wu et al., 2020). The alphacoronaviruses and betacoronaviruses infect only mammals; gammacoronaviruses and deltacoronaviruses infect birds, but some of them can also infect mammals. The genome of coronaviruses is arranged in the order of the 5′ untranslated region (5′ UTR), open reading frame 1a/b (orf1ab), spike (S) protein, envelope (E) protein, membrane (M) protein, nucleoprotein (N) protein, 3′ UTR, and the poly (A) tail, with regions encoding accessory proteins, including orf3, 6, 7a, 7b, 8, and 9b, between those encoding structural proteins (Fig. 1B). The large replicase polyproteins pp1a and pp1ab, encoded by the partially overlapping 5′-terminal orf1a/b within the 5′ two-thirds of the genome, are proteolytically cleaved into 16 putative nonstructural proteins (nsps; Fig. 1B).
Fig. 1.
The genomic structure of coronaviruses. (A) Classification of coronavirus, with the new coronavirus SARS-CoV-2 highlighted in red. (B) The genome structure of four genera of coronaviruses. Pp1a and pp1b represent the 2 long polypeptides that are processed into 16 nonstructural proteins. S, E, M, and N indicate the four structural proteins spike, envelope, membrane, and nucleocapsid proteins.
In early December 2019, a cluster of cases of pneumonia caused by a novel coronavirus named Sudden Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) resulted in tremendous challenges to China's public health and clinical treatment (Munster et al., 2020; Yan et al., 2020), and now it has been confirmed in more than 211 other countries and territories, causing a major global public health crisis (Day, 2020; Jernigan and Team, 2020; Peeri et al., 2020). SARS-CoV-2 belongs to the Betacoronavirus genus in the family Coronaviridae, and is closely related to severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) (Drosten et al., 2003; Wassenaar and Zou, 2020; Zaki et al., 2012). SARS-CoV, MERS-CoV and SARS-CoV-2 are three highly transmissible and pathogenic viruses that have emerged in humans in the past 2 decades and bats are considered their natural hosts (Zhou et al., 2020). They mainly infect the lower respiratory tract and cause severe pneumonia (Hotez et al., 2020), sometimes leading to fatal acute lung injury and acute respiratory distress syndrome, resulting in high morbidity and mortality. Cytokines and chemokines, which are triggered by the innate immune system during virus infections have long been thought to play an important role in immunity, but dysregulated and excessive immune responses may cause immunopathology (Channappanavar and Perlman, 2017). Research shows that robust SARS-CoV replication accompanied by delayed IFN-I signaling promotes the accumulation of pathogenic inflammatory monocyte-macrophages, resulting in elevated lung cytokine/chemokine levels, vascular leakage, and impaired virus-specific T cell responses (Channappanavar et al., 2016). In addition to SARS-CoV and MERS-CoV, SARS-CoV-2 infection also can induce proinflammatory cytokine responses, including interferon gamma (IFN-γ), tumor necrosis factor alpha (TNFα), interleukin-1 (IL-1), IL-2, IL-6 and IL-8 (Kadkhoda, 2020). In patients with severe COVID-19, lymphopenia is a common feature, with drastically reduced numbers of CD4+ T cells, CD8+ T cells, B cells and natural killer (NK) cells, as well as a reduced percentage of monocytes, eosinophils and basophils. Furthermore, SARS-CoV-2 may have an antibody-dependent enhancement effect (Cao, 2020; Xu et al., 2020).
Although there have been outbreaks of coronavirus diseases, such as SARS and MERS, when the novel coronavirus SARS-CoV-2 outbreak occurred, there were no effective vaccines or drugs available to prevent or cure the disease. Clinical development of multiple vaccines and screening of specific antiviral drugs were initiated in parallel with attempts to contain the outbreak. Currently, more than 115 candidate vaccines for SARS-CoV-2 are being developed globally. Five advanced candidates have recently moved into clinical development, including mRNA-1273 from Moderna, Ad5-nCoV from CanSino Biologicals, INO-4800 from Inovio, LV-SMENP-DC and pathogen-specific aAPC from Shenzhen Geno-Immune Medical Institute (Thanh Le et al., 2020).
It became apparent that coronaviruses can cross the species barrier and cause life-threatening infections in humans (Ji et al., 2020; Nishiura et al., 2020). Therefore, further attention needs to be paid to these new coronaviruses. In addition, coronaviruses causing high pathogenicity and high mortality in animals have attracted increasing attention. For example, highly virulent porcine epidemic diarrhea virus (PEDV), which appeared in China in 2010 and the United States in 2013, caused large global economic losses to the pig industry due to the lack of effective vaccines and drugs (Kong et al., 2019; Li et al., 2012; Stevenson et al., 2013). Another coronavirus, swine acute diarrhea syndrome coronavirus (SADS-CoV), which is a novel bat-HKU2-like coronavirus, was responsible for a large-scale fatal outbreak of in pigs in China that caused the death of more than 24,000 piglets across four farms (Zhou et al., 2018). Obviously, the role of vaccines is an effective way not only to enhance the ability of individuals to fight the virus but also to stop the spread of the virus. To be better prepared for future outbreaks of unknown human coronavirus pathogens, platform technologies to accelerate vaccine development should be employed.
2. Coronavirus reverse genetics systems
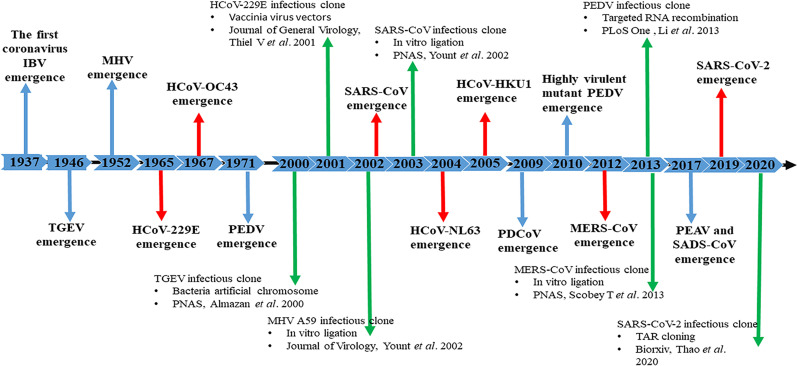
Reverse genetics systems are highly valuable research tools for RNA virus vaccine development, which may be more efficient than conventional approaches with live attenuation through passing (forward genetics) or inactivation (Stobart and Moore, 2014). Reverse genetics systems are useful tools for studying the modifications of viral genomes and for generating recombinant viruses to better understand their fundamental biology, develop novel vaccine candidates, and test antiviral therapeutics. Recently, reverse genetics techniques, including targeted RNA recombination, in vitro ligation and bacterial artificial chromosome systems, vaccinia virus vectors and transformation associated recombination (TAR) cloning, have been successfully used to manipulate the genome of coronaviruses (Fig. 2 ).
Fig. 2.
Timeline of emerging representative coronavirus events and their infectious clones generated using reverse genetics systems. The timeline spans from the first coronavirus in history to the emergence of 2019-nCoV in 2019. Red arrows indicate human coronavirus outbreaks and identification events. Blue arrows indicate animal coronavirus outbreaks and identification events. Green arrows indicate the publication of infectious clones using different reverse genetics methods (Almazan et al., 2000; Li et al., 2013; Thao et al., 2020; Yount et al., 2002).
2.1. Reverse genetic system using targeted RNA recombination
Targeted RNA recombination was the first reverse genetic system devised for coronaviruses at a time when it was not clear whether a full-length clone could be successfully constructed (Masters and Rottier, 2005). Constructing an infectious clone of a coronavirus using targeted RNA recombination requires two steps: first, constructing a chimeric coronavirus that carries the mouse hepatitis virus (MHV) S gene and has a strict mouse cell affinity and second, rescuing the targeted recombinant coronavirus on a specific cell dependent on the virus (Fig. 3A). Targeted RNA recombination was the first reverse genetic system devised for MHV. Subsequently, the method was applied to the construction of full-length cDNA in other coronaviruses, including PEDV and feline infectious peritonitis virus (FIPV) (Haijema et al., 2003; Li et al., 2013). Targeted RNA recombination presents clear limitations; for example, it does not operate on parts of the genome that code for replicating enzymes.
Fig. 3.
Flowchart of three methods for constructing coronavirus infectious cloning. (A) Targeted RNA recombination scheme used to make interspecies chimeric viruses: mIBV (Stage 1) and recombinant IBV (Stage 2). (B) In vitro ligation. The SARS-CoV full-length genome is divided into seven segments, named A-F, with type II restriction endonucleases Bgl1 at both ends. (C) Bacterial artificial chromosome system. Restriction enzyme sites in the MERS-CoV genome were employed to ligate the full-length MERS-CoV genome into pBeloBAC11.
2.2. Reverse genetic system using in vitro ligation
In vitro ligation uses unique type II restriction endonucleases (e.g., BglI, SapI, and BsaI) that cleave several bases away from their recognition site, allowing for the reassembly of authentic coronavirus genomes from smaller fragments. Detailed steps can be found in the published literature (Cockrell et al., 2017). The assembled full-length cDNA, containing a T7 RNA polymerase promoter at the 5′ end and a poly (A) tail at the 3′ end, is transcribed in vitro to generate capped full-length transcripts that are used together with capped N gene transcripts to efficiently rescue infectious virus after the transfection of susceptible cells (Fig. 3B). The first coronavirus full-length infectious cDNA clone was also generated for TGEV using in vitro ligation (Yount et al., 2000). In addition, SARS-CoV, MERS-CoV, PEDV and SARS-CoV-2 were constructed using in vitro ligation (Scobey et al., 2013; Xie et al., 2020; Yount et al., 2003; Zaki et al., 2012). This method can avoid the instability and virulence of viral cDNA in bacteria, as it depends on the T7 RNA polymerase.
2.3. Reverse genetic system using bacteria artificial chromosomes
The bacterial artificial chromosome (BAC) system allows the assembly of a full-length cDNA copy of the viral genome in BAC plasmids such as pBeloBAC11, a low-copy-number plasmid that presents strictly controlled replication, leading to one or two plasmid copies per cell (Almazan et al., 2008). BAC plasmids contain a 5′ cytomegalovirus promoter of the viral genome, allowing for the transcription of the viral genome following the transfection of BAC DNA into mammalian cells. In addition, coronavirus BACs contain a poly (A) tail, a hepatitis D virus ribozyme, and bovine growth hormone termination and polyadenylation signals to create genomic RNA with an authentic 3′ end (Fig. 3C). The first full-length infectious coronavirus cDNA clone was generated for transmissible gastroenteritis virus (TGEV) using the BAC system. In addition, SARS-CoV, MERS-CoV, FIPV and PEDV have been successfully developed (Balint et al., 2012; Li et al., 2017). The BAC system presents several advantages, including the high stability of exogenous sequences, unlimited production of the cDNA clone, high efficiency of cDNA transfection into mammalian cells, easy manipulation for genes and intracellular expression of the viral RNA (Fehr, 2020).
2.4. Reverse genetic system using transformation associated recombination cloning
Recently, transformation associated recombination (TAR) cloning has been used to construct infectious clones of coronaviruses, including MHV, MERS-CoV and SARS-CoV-2 (https://www.biorxiv.org/content/10.1101/2020.02.21.959817v1). The overall strategy is as follows (Fig. 4A): viral RNA is prepared from several overlapping DNA fragments by RT-PCR or chemical synthesis (no virus available). The 5′- and 3′-terminal DNA fragments contain the T7-RNA polymerase promoter upstream of the coronavirus at the 5′ end, a restriction endonuclease cleavage site downstream of the poly(A) sequence, and overlapping sequences with the TAR plasmid pVC604. Then, all the fragments are used to transform Saccharomyces cerevisiae (strain VL6-48N), and the positive colonies are screened for the correct assembly of the yeast artificial chromosome (YAC) containing virus genome and purified the YAC. The plasmid is linearized using restriction endonuclease and subjected to T7 RNA polymerase-based in vitro transcription to generate transcripts. The RNA transcripts together with capped N gene transcripts are transfected into BHK cells to obtain the corresponding virions. The TAR cloning system can rapidly reassemble at least 14 fragments with remarkable efficacy (usually > 90% of the clones are correct).
Fig. 4.
Flowchart of two methods for constructing coronavirus infectious clones. (A) Transformation-associated recombination (TAR) cloning. A schematic representation of the general workflow of TAR cloning for SARS-CoV-2 infectious clones. (B) Vaccinia virus vectors. The general workflow of vaccinia virus vectors for constructing HCoV-229E infectious clones.
2.5. Reverse genetic system using vaccinia virus vectors
The vaccinia virus cloning vector also can be used to construct infectious coronavirus clones and represents a generic approach to reverse genetics (Thiel and Siddell, 2005). Vaccinia virus vectors allow large fragments (26–31 kb) of foreign genes to be inserted without affecting virus replication, and vaccinia viruses can easily produce high titer strains. The process of cloning coronavirus cDNA in vaccinia virus is as follows (Fig. 4B). First of all, the virus genome is analyzed for useful naturally encoded endonuclease restriction sites that can later be used to ligate cloned cDNA inserts and generate a set of plasmid DNAs that cover the full-length virus genomic sequence. Upstream of the viral 5′ end, there should be an EagI or a Bsp120I restriction endonuclease site to allow insertion of the cDNA into the NotI site of the vaccinia virus genomic DNA by in vitro ligation. Downstream of the 3′ end of the virus genome, a poly(A) tail and unique restriction endonuclease site should be cloned. Second, long cDNA fragments are assembled by in vitro ligation to obtain a full-length virus cDNA fragment. Third, the full-length fragment or several fragments are inserted into a vaccinia virus genome by in vitro ligation. Finally, the vaccinia virus containing the full-length cDNA virus genome is linearized and used as a template for in vitro transcription using bacteriophage T7 RNA polymerase and transfected into BHK cells to obtain the corresponding virions. Currently, vaccinia virus vectors are successfully applied to reverse genetic manipulation of several coronaviruses, including HCoV-229E, IBV, MHV and SARS-CoV (Thiel et al., 2001; Thiel and Siddell, 2005; van den Worm et al., 2012).
2.6. Rapid gene manipulation of coronavirus
Chemical synthesis of the viral genome is widely used for the construction of infectious clones. Using a chemical synthesis system, the complete genome of the virus can be synthesized to construct a full-length genomic cDNA clone, especially in the absence of a natural template. This method was recently used to construct an infectious clone of SARS-like CoV (Becker et al., 2008). In general, the ability to quickly and efficiently introduce virtually any type of genetic modification (point mutations, insertions, or deletions) into the coronavirus genome and recover recombinant virus is desirable. CRISPR/Cas9 technology is an efficient platform for manipulating the modification of full-length infectious cloning of coronaviruses, which can generate recombinant viruses within a week (Peng et al., 2020; Shen et al., 2019; Wang et al., 2019a). Moreover, a combination of bacterial artificial chromosome and lambda red recombination with the I-SceI homing endonuclease method was used to engineer genetic alterations of the MERS-CoV (Fehr, 2020; Muth et al., 2017). Importantly, the optimized reverse genetics platform used in this study will simplify the construction of mutant infectious clones and help accelerate the progress in coronavirus research.
3. Rational design of vaccine candidates using reverse genetics technology
Vaccination is expected to be an efficacious strategy in preventing individuals and animals from suffering from coronavirus infections. Virus vaccines must be immunogenic and sufficiently stable, safe, and suitable to induce long-lasting immunity. To date, various kinds of candidate vaccines for coronaviruses have been developed, including live attenuated vaccines, subunit vaccines, DNA vaccines, inactivated vaccines, and recombinant vector vaccines. Each vaccine type has different advantages and disadvantages. For example, traditional inactivated vaccines can generally induce highly potent immune responses and/or protection, but they may induce the antibody-dependent enhancement effect. In addition, they exit recovery of virulence if incomplete inactivation of viruses, resulting in significant safety concerns (Luo et al., 2018). Although DNA vaccines maintain strong safety, they are generally less immunogenic than are whole-virus-based vaccines, usually requiring the optimization of sequences or immune routes; and adjuvant coordination is also often required to achieve immune effects. Among them, live attenuated vaccines are considered highly effective because of their ability to replicate within host cells, inducing high levels of antigenic stimulation and robust long-term immunological memory. However, a major safety concern with live attenuated vaccines is the possibility of reversion to a pathogenic form and oral or aerosol route vaccination that can induce mucosal immune responses should be considered.
3.1. Live attenuated vaccines
Virus-infected cells react quickly to invading viruses by producing type I interferons (IFN-α/β) and type III interferons (IFN-λ1/λ2/λ3/λ4) and establish an innate antiviral state, which provides a first line of defense against viral infection. Type III IFNs are mainly produced by epithelial cells and play a major role in diseases that cause mucosal immunity, such as PEDV and PDCoV. However, most coronaviruses commonly suppress interferon levels in infected cells. Reverse genetics technology provides a very useful platform in the study of virulence and host innate immune-related mutations and in the rational design of coronavirus live attenuated vaccine candidates. According to current research, the 5′ UTR, several structural proteins, nonstructural proteins and the 3′ UTR of coronaviruses all have more or less influence on virulence and the life cycle of the virus. Here, we summarize the coronavirus genes associated with virulence, replication and innate immunity (Table 1 ) and potential vaccines based on reverse genetics (Table 2 ).
Table 1.
Summary of different proteins (nonstructural, structural, and accessory proteins) in coronavirus pathobiology.
| Classification | Protein | Pathobiology | Reference |
|---|---|---|---|
| Nonstructural proteins | nsp1 | A vital virulence factor; inhibits the expression of the host gene; inhibits IFN production | Shen et al. (2019) and Zhang et al. (2018b) |
| nsp2 | Related to viral replication; regulates inflammation via NF-κB activation | Graham et al. (2005) and Wang et al. (2018b) | |
| nsp3 | An essential component of the replication/transcription complex; inhibits IFN production | Lei et al. (2018) | |
| nsp4 | Related to cellular membrane rearrangements and viral replication | Beachboard et al. (2015) | |
| nsp5 | Viral replication; inhibits IFN production; cleaves polypeptides | Chen et al. (2019) | |
| nsp6 | Related to autophagy and cellular membrane rearrangements | Angelini et al. (2013) and Cottam et al. (2014) | |
| nsp7 | Inhibits IFN production (PEDV) | Zhang et al. (2016) | |
| nsp8 | Inhibits IFN production (PEDV) | Zhang et al. (2018b) | |
| nsp9 | Dimerization and RNA binding | Egloff et al. (2004) | |
| nsp10 | Interacts with both nsp14 and nsp16; regulates SARS-CoV replication function and replication fidelity | Bouvet et al. (2014) and Smith et al. (2015) | |
| nsp14 | Has 3′-to-5′ exoribonuclease activity and N7-methyltransferase (N7-MTase) activities; vital for virulence; inhibits IFN production | Case et al. (2018) and Graepel et al. (2017) | |
| nsp15 | Endoribonuclease; vital virulence factor | Deng et al. (2017) and Zhang et al. (2018a) | |
| nsp16 | Has 2′-O-methyltransferase (MTase) activity; vital virulence factor; negatively regulates innate immunity | Hou et al. (2019b) and Yong et al. (2019) | |
| Structural proteins | S | Receptor binding and membrane fusion; vital virulence factor; target protein of a subunit vaccine | Hou et al. (2019a) and Shang et al. (2020a) |
| M | Morphogenesis or assembly virus; inhibits IFN production | Lui et al. (2016) | |
| E | Involved in the viral life cycle, including assembly, budding, and pathogenesis; vital virulence factor; ion channel | Schoeman and Fielding (2019) and Shang et al. (2020b) | |
| N | Regulates viral RNA synthesis, packaging of the viral RNA in helical nucleocapsids and in virion assembly; inhibits IFN production | Lu et al. (2011) and Veit et al. (2018) | |
| Accessory proteins | ORF3 | Virulence factor; dispensable; inhibits IFN production; ion channel | Menachery et al. (2017b) and Van Beurden et al. (2018) |
| ORF4 | Virulence factor; dispensable | Menachery et al. (2017b) | |
| ORF5 | Virulence factor; dispensable | Menachery et al. (2017b) and Van Beurden et al. (2018) | |
| ORF6 | Inhibits IFN production | Hu et al. (2017) and Kopecky-Bromberg et al. (2007) | |
| NS6 | Virulence factor; dispensable; inhibits IFN production | Fang et al. (2018) | |
| ORF8a | Virulence factor; dispensable | Castano-Rodriguez et al. (2018) |
Table 2.
Summary of the application of reverse genetics in live attenuated vaccines of different members of Coronaviridae.
| Coronaviridae | Virus | Mutation sites | Evaluation | Reference |
|---|---|---|---|---|
| Alphacoronavirus | PEDV | nsp15: H262A | Attenuation | Deng et al. (2019) |
| nsp16: KDKE/AAAA | Attenuation and protective immunity against parental strain infection | Hou et al. (2019a) | ||
| S: ΔYxxΦEKVHVQ of the S protein or Δ197aa | ΔYxxΦEKVHVQ: attenuation Δ197aa: attenuation, reduces the protection |
Hou et al., 2017, Hou et al., 2019b | ||
| ORF3: ΔORF3 | Attenuation | Beall et al. (2016) | ||
| Betacoronavirus | MERS-CoV | Deletion: ΔE,3, 4a, 4b, and 5 | None | Almazan et al. (2013) |
| nsp16: D130A | Attenuation and protection against a lethal MERS-CoV challenge | Menachery et al. (2017a) | ||
| MHV | nsp1: Δ99 nts (829–927 nt) or Δ27 nts between nts 780–808 (LLRKxGxKG) | Δ99 nts: attenuation and protection of mice from homologous and heterologous viral infections Δ27 nts: attenuation and protection against challenge with WT MHV-A59 virus |
Lei et al. (2013) and Zust et al. (2007) | |
| nsp2: Δnsp2 | None | Graham et al. (2005) | ||
| nsp3: V787S or N1347A | V787S: reduced pathogenesis and protection against challenge with WT virus N1347A: reductions in lethality and weight loss |
Fehr et al. (2015) and Mielech et al. (2015) | ||
| nsp5: T26I/D65G | Attenuation | Deng et al. (2014) | ||
| nsp15: H262A | Attenuation and protective immunity against WT virus infection | Deng et al. (2017) | ||
| SARS-CoV | nsp1: ΔLLRKNGNKG (121–129aa)ΔEDYEQNWNTKH(154–164aa) | No significant weight loss and 100% survived the challenge | Jimenez-Guardeno et al. (2015) | |
| nsp2: Δnsp2 | None | Graham et al. (2005) | ||
| nsp3: N1040A | Attenuation | Fehr et al. (2016) | ||
| nsp14: D90A/E92A | Reductions in weight loss and lung titer | Graham et al. (2012) | ||
| nsp16: D130A | Attenuation in a variety of pathogenic outcomes and minimal weight loss | Menachery et al. (2014) | ||
| E: ΔPBM of the E protein | Decreased lung pathology | Jimenez-Guardeno et al. (2014) | ||
| Gammacoronavirus | IBV | Δ3ab/5ab | Attenuation and protection against a homologous challenge | Van Beurden et al. (2018) |
| Δ3ab/5ab of Beaudette and the S gene was replaced by the S gene from the M14 strain | Attenuation and protection against a M14 strain challenge | Van Beurden et al. (2018) | ||
| Deltacoronavirus | PDCoV | NS6:ΔNS6 | Attenuation | Zhang et al. (2020) |
It is well known that among four coronavirus genera, only alphacoronaviruses and betacoronaviruses produce the nsp1 protein, which is located at the N-terminal of the replicase polyprotein pp1a. In terms of both sequence identity and size, nsp1 of alphacoronaviruses shares low similarity with nsp1 of betacoronaviruses. To date, the crystal structure of nsp1 from several coronaviruses (SARS-CoV, TGEV, and PEDV) has been resolved, which is important to understand the role of nsp1 in the pathogenesis of viruses and its interaction with host cell proteins (Shen et al., 2018, Shen et al., 2019). The nsp1 protein of coronaviruses is a vital virulence factor widely demonstrated in both alphacoronaviruses and betacoronaviruses (Jimenez-Guardeno et al., 2015; Lokugamage et al., 2015; Shen et al., 2018, Shen et al., 2019; Zhang et al., 2015). Not only inhibiting the expression of host genes with different mechanisms, but also playing a significant regulatory role in evading the host's natural immune response, nsp1 mainly inhibits the production of IFNs (Zhang et al., 2016, Zhang et al., 2018c). Additionally, some studies have shown that abolishing the anti-IFN function of nsp1 can attenuate viruses, which has been demonstrated in MHV and SARS-CoV (Jimenez-Guardeno et al., 2015; Lei et al., 2013).
The nsp2 proteins of some coronaviruses are dispensable for viral replication in cell culture, but those viruses lacking nsp2, such as MHV and SARS-CoV, show attenuated viral growth and RNA synthesis (Graham et al., 2005). However, the pathogenicity of these viruses in mice has not been further reported. In addition, amino acids 1–120 of TGEV nsp2 are involved in the regulation of inflammation via NF-κB activation (Wang et al., 2018b).
The multidomain in nsp3, an essential component of the replication/transcription complex, is the largest protein encoded by the coronavirus genome, with an average molecular mass of approximately 200 kDa (Lei et al., 2018). In all coronaviruses, highly conserved functional domains within nsp3 are present: papain-like protease domains, ubiquitin-like domains, and ADP-ribose-phosphatase domains (Lei et al., 2018). Using a recombinant SARS-CoV strain with reduced nsp3 de-ADP-ribosylation activity showed that this mutant strain led to virus attenuation in mice but protected them from an otherwise lethal SARS-CoV infection and significantly enhanced the innate immune response, indicating that it is an important virulence factor for SARS-CoV (Fehr et al., 2016). V787S of nsp3 is thought to induce attenuation of MHV, and the mutant MHV strain elicits protective immunity (Mielech et al., 2015). N1347A of nsp3 is also thought to be associated with the virulence of MHV (Fehr et al., 2015). PEDV and TGEV (590–1215aa, via NF-κB) nsp3 proteins have been proven to evade the host immune system (Wang et al., 2019b). In addition, 102 and 61 amino acid substitutions were found in nsp2 and nsp3 of SARS-CoV-2 compared with SARS-CoV or SARS-like bat CoV, respectively, and these differences warrant further investigation (Wu et al., 2020).
The nsp4 genes of coronaviruses are significant in viral replication and pertain to cellular membrane rearrangements (Beachboard et al., 2015). Both H120 and F121 in SARS-CoV nsp4 play critical roles in viral replication (Sakai et al., 2017). However, whether nsp4 affects the virulence of the virus needs to be further verified by using reverse genetics.
The coronavirus nsp5 protease (3CLpro; Mpro) processes nsp proteins at 11 cleavage sites and is essential for virus replication, making it a high value target for the development of anti-coronavirus therapeutics (Jo et al., 2020; Tomar et al., 2015). It has been reported that coronavirus nsp5 cleaves NF-κB essential modulator and STAT2, inhibiting IFN production, which has been verified in FIPV, PEDV and PDCoV (Chen et al., 2019; Wang et al., 2016; Zhu et al., 2017). MHV nsp5 T26I/D65G, which confers resistance to a broad-spectrum coronavirus 3C-like protease inhibitor, also has a slight effect on the virulence and replication of MHV (Deng et al., 2014).
Coronavirus nsp6 is associated with autophagy (Cottam et al., 2014). Scientists used reverse genetics to verify that deletion of any of regions encoding nsp7, nsp8, nsp9 and nsp10 in MHV was lethal to virus (Deming et al., 2006). Furthermore, PEDV nsp7 was found to inhibit the IFN-β and IRF3 promoter activities (Zhang et al., 2016), and nsp8 can suppress type III IFN activities (Zhang et al., 2018b).
The 148-amino acid nsp10 subunit contains 2 zinc fingers and is known to interact with both nsp14 and nsp16, stimulating their respective 3′-5′ exoribonuclease and 2′-O-methyltransferase activities. A series of studies have shown that nsp10 is a major regulator of the SARS-CoV replication function (Bouvet et al., 2014). The single and double mutations R80A/E82A-ExoN(+) in MHV nsp10 disrupt the nsp10–nsp14 interaction, and compared to the wild-type virus ExoN(+), these mutations rendered the virus 5 and 10 times more sensitive to treatment with the RNA mutagen 5-fluorouracil, respectively. The results showed that nsp10 is important for CoV replication fidelity and support the hypothesis that nsp10 functions to regulate nsp14-ExoN activity during viral replication (Smith et al., 2015).
Coronavirus nsp14 has 3′-to-5′ exoribonuclease (ExoN) and N7-methyltransferase (N7-MTase) activities. The former is a proofreading function that is required for high-fidelity replication. The inactivation of MHV-CoV ExoN(−) activity by the nsp14 mutations D89A and E91A are relatively stable, even after passaging MHV-ExoN(−) 250 times, without reversion of the ExoN(−) mutation site. However, novel amino acid changes within the RNA-dependent RNA polymerase and nsp14 of MHV-ExoN(−) P250, indicate that multiple replicase proteins could compensate for the ExoN function during replication and will likely inform the design of countermeasures for endemic and emerging CoVs by defining novel common targets for stable virus attenuation or direct inhibition (Graepel et al., 2017). MHV-ExoN activity is required for resistance to the innate immune response (Case et al., 2018). The inactivation of SARS-CoV ExoN activity by the nsp14 mutations D90A and E92A was attenuated in both young and aged disease models compared to virulent WT, and it provided complete protection against lethal challenge in a susceptible, immunosenescent mouse model of the lethal SARS-CoV infection (Graham et al., 2012). The recombinant TGEV virus with a mutation in zinc finger 1 of the ExoN domain of nsp14 induced weak antiviral responses, including reduced expression of beta interferon (IFN-β), tumor necrosis factor (TNF), and interferon-stimulated genes. Therefore, coronavirus nsp14 plays a potential role in regulating innate immune responses (Becares et al., 2016). In addition, PEDV nsp14 was found to suppress type I and type III IFN activities (Zhang et al., 2016, Zhang et al., 2018c).
Nsp15 encoded by coronavirus is a nidoviral uridylate-specific endoribonuclease (NendoU) that plays an essential role in the life cycle of the virus (Zhang et al., 2018a). MHV with the nsp15 H262A mutation that inactivates the EndoU activity of nsp15, resulted in greatly attenuated disease in mice and stimulated a protective immune response (Deng et al., 2017). PEDV EndoU activity is also a key virulence factor and coronavirus nsp15 is a target for generating live attenuated vaccine (Deng et al., 2019).
Nsp16 has 2′-O-methyltransferase (MTase) activity and it is conserved across the entire coronavirus family, increasing its appeal as an attenuation target (Yong et al., 2019). Several recent works have shown that although disruption of nsp16 activity in MHV (D129A), SARS-CoV (D130A), MERS-CoV (D130A) and PEDV (KDKE to AAAA) rendered an attenuated strain, it induced stronger type I and type III interferon responses and protected animals from the challenge (Hou et al., 2019a; Menachery et al., 2017a, Menachery et al., 2018). However, without sufficiently attenuated virulence, the nsp16 mutant could be combined with another attenuating mutation in nsp1, nsp14, or S protein, which will produce a stable, attenuated virus capable of protection from heterologous challenge (Hou et al., 2019a; Menachery et al., 2018).
The coronavirus S protein, which is a class I fusion protein located at the surface of the virus, includes the receptor binding S1 subunit and the membrane fusion S2 subunit (Shang et al., 2020a; Walls et al., 2020). The coronavirus S protein is considered a major target for the development of subunit vaccines and major virulence genes. Two chimeric viruses with the reciprocally exchanged S gene were generated; one is a highly virulent strain, and the other is an avirulent strain, showing that the S gene is only one of the necessary determinants for virus virulence (Wang et al., 2018a). Another report examined the role of the S gene in PEDV pathogenesis with different generations of strains in the same way. Although the results showed that the S gene plays a role in virulence, other genes might also important in determining virulence (Kao and Chang, 2019). Three recombinant IBVs, BeauR-M41 (S1), BeauR-QX (S1) and BeauR-M41 (S), based on the BeauR backbone expressing a heterologous S1 or S from M41 or QX conferred incomplete protection against homologous challenge based on ciliary activity and clinical signs. However, the protection efficiency of BeauR-M41(S) was higher than that of BeauR-M41 (S1) (Ellis et al., 2018). Coronavirus virions assemble at the endoplasmic reticulum (ER)-Golgi intermediate compartment (ERGIC) and some of them, including SARS-CoV, TGEV, IBV, and PEDV, possess conserved motifs of YxxΦ and/or KxHxx/KKxx in the cytoplasmic tail of the S protein (Hou et al., 2019b; Ujike et al., 2016). A recombinant PEDV with deletions or a mutation in the two motifs (YxxΦ and/or KxHxx/KKxx) showed that the YxxΦ motif triggers endocytosis of S proteins and that the motif KVHVQ is involved in the retention of the S proteins in the ER-Golgi intermediate compartment; the loss of both motifs significantly enhances syncytia formation in Vero cells and reduces virulence in pigs (Hou et al., 2019b). The NTD of the S protein can attenuate a highly virulent coronavirus, which was confirmed by a recombinant virus (icPC22A-S1Δ197aa) and (icTGEV-S1Δ224aa), but the virus may lose important epitopes for inducing robust protective immunity, which provides new insights for the development of a new attenuated vaccine (Hou et al., 2017; Wang et al., 2019a). In addition, 27 amino acid substitutions were found in the SARS-CoV S protein compared with SARS or SARS-like CoV, and whether these differences could affect the host tropism and transmission property is worthy of future investigation (Wu et al., 2020).
The coronavirus E protein is the smallest protein of the major structural proteins and is an integral membrane protein involved in several aspects of the virus life cycle, including assembly, budding, and pathogenesis (Schoeman and Fielding, 2019). Viruses lacking E protein form infectious virions, but their replication efficiency and virulence are significantly reduced. This seems to indicate that the E protein is an important virulence factor in coronavirus but is not essential for some coronaviruses (Castano-Rodriguez et al., 2018; Fett et al., 2013; Jimenez-Guardeno et al., 2015; Shang et al., 2020b). MERS-CoV and TGEV lacking the E gene are not successfully rescued, but they can spread from cell to cell via expression of the E protein (Almazan et al., 2013; Ortego et al., 2002).
Coronavirus M proteins are major structural proteins and are required for virion assembly. The S, E and M proteins can form virus-like particles (VLPs) with structural integrity (Wang et al., 2017). Both peptides (M1–31 and M132–161) in the SARS-CoV M protein are immunogenic (He et al., 2005), but the M protein of SARS-CoV has not been investigated for its protective efficacy against SARS-CoV infection. Several studies have shown that the coronavirus M protein also inhibits IFN production (Shokri et al., 2019; Siu et al., 2009). The MERS-CoV M protein inhibits type I IFN production through the inhibition of TBK1-dependent phosphorylation of IRF3 (Lui et al., 2016).
Coronavirus N proteins have been associated with multiple functions in the viral life cycle, including the regulation of viral RNA synthesis, the packaging of the viral RNA in helical nucleocapsids, and virion assembly through the interaction with the viral M protein (Mcbride et al., 2014). Expression of the N protein is also necessary for efficient recovery of the virus from infectious cDNA clones. Unlike the S protein, the N protein has no ability to elicit neutralizing antibodies, but it may induce specific antibody and cellular immune responses. The N protein plays an important role in viral pathogenesis since BALB/c mice immunized with recombinant virus MVA-MERS-N exhibit stronger T cell responses and anti-N monoclonal antibodies protect mice from lethal infection by MHV (Nakanaga et al., 1986; Veit et al., 2018). Immunodominant SARS-CoV-N regions N1 (1–422aa) and N3 (110–422aa) produce specific antigens in BALB/c mice and react with the serum of SARS patients; hence, they can be used as effective SARS DNA vaccines (Dutta et al., 2008). The SARS-CoV N protein is also an IFN antagonist that inhibits IFN synthesis by inhibiting IRF-3 and NF-κB, interfering with TRIM25-mediated RIG-I ubiquitination or attenuating PTAC-mediated RIG-I/MDA5 activation (Ding et al., 2017; Hu et al., 2017).
Coronavirus accessory proteins have been generally regarded as dispensable for in vitro viral replication. For example, PEDV with the ORF3 gene entirely deleted could replicate similarly to wild-type PEDV (Beall et al., 2016). Likewise, deletions of NS6 from PDCoV, ORF8a from SARS-CoV, ORF3, ORF4 and ORF5 from MERS-CoV and ORF3 and ORF5 from IBV do not affect replication, and the deletion of these accessory proteins could attenuate viruses, showing that accessory proteins are also related to the virulence of viruses (Almazan et al., 2013; Castano-Rodriguez et al., 2018; Menachery et al., 2017b; Zhang et al., 2020). The accessory proteins of coronaviruses are often used as target regions for the insertion of exogenous gene since they are dispensable. Notably, some accessory proteins have ion channels and are related to the innate immune response (Kint et al., 2016; Wong et al., 2018; Yue et al., 2018; Zeng et al., 2016). Some accessory proteins interact with host cells, potentially helping viruses evade the immune system and increase their virulence (Comar et al., 2019; Fang et al., 2018). ORF4a, ORF4b and ORF8b of MERS-CoV seem to be involved in viral evasion of NF-κB mediated host immune responses (Lee et al., 2019). Therefore, the mutant virus lacking some accessory proteins is potentially a safe and promising vaccine candidate to prevent coronavirus infection.
3.2. Recombinant vector vaccines
To date, viral vectors utilizing modified vaccinia virus Ankara, measles virus, RV vectors or adenovirus have been used to express the glycoprotein of coronaviruses, including SARS-CoV and SARS-CoV-2, and have been shown to be immunogenic according to in vivo assays (Kato et al., 2019; Thanh Le et al., 2020). To date, a recombinant novel coronavirus vaccine using an adenovirus type 5 vector that expresses the S protein has entered phase I clinical trials (Thanh Le et al., 2020). Researchers designed recombinant replication-deficient adenovirus-based vaccines expressing the MERS-CoV S protein that induced the highest neutralizing antibody titer and the strongest cytokine-induced T cell responses, and these may confer protection against MERS-CoV infection (Kim et al., 2019). Recombinant parainfluenza virus 5 (PIV5)-expressing the MERS-CoV S protein can induce neutralizing antibodies and robust T cells in a human DPP4 knock-in C57BL/6 congenic mouse model (hDPP4 KI) and protect mice from fatal MERS-CoV infection (Li et al., 2020b). An attenuated parainfluenza virus encoding the full-length S protein of SARS-CoV was used for the vaccination of African green monkeys, and monkeys with this vaccine were protected from subsequent homologous SARS-CoV infection (Bukreyev et al., 2004). Another novel recombinant influenza A virus (H1N1pdm09) with MERS-CoV was generated, and the inactivated chimeric bivalent vaccine induced potent and specific neutralizing antibodies against MERS-CoV and H1N1pdm09 in BALB/c mice (Shehata et al., 2019). The use of reverse genetics to construct recombinant vaccine strains could achieve a single immunization and prevent two diseases.
4. Study the cell and tissue tropism of coronaviruses using reverse genetics
Coronaviruses generally exhibit restricted cell and tissue tropism, which is dependent on the S glycoprotein of individual coronavirus strains (Millet and Whittaker, 2015). Understanding the cell and tissue tropism of a virus is helpful to further research the pathogenesis of the virus and save vaccine production costs. Vaccines against IBV, both live attenuated and inactivated, are currently produced in embryonated hen eggs, which is a cumbersome and expensive process, because most IBV strains do not replicate in cultured cells. Previous studies have shown that the S2 subunit of the avirulent BeauR strain is responsible for its extended cellular tropism for Vero cells (Bickerton et al., 2018b). Later, recombinant IBVs with the immunogenic S1 subunit were derived from the IBV vaccine strain and the virulent field strain was generated using reverse genetics. The recombinant IBVs are able to replicate in both primary chicken kidney and Vero cells, which would enable IBV vaccines to be grown in cell lines rather than expensive embryonated hen eggs (Bickerton et al., 2018a). Moreover, recombinant IBV with a mutant S2′ site (furin S2′ site) led to neurotropism (Cheng et al., 2019). TGEV has both enteric and respiratory tropism. TGEV S219A in the S protein was required to confer enteric tropism, and a 6 nt insertion at position 1124 could increase virus stability and virus titers after passage in cell cultures (Sanchez et al., 2019). The MHV amino acid lysine at 194 in the nsp1 protein can promote virus replication in the liver, and this study further confirmed that nsp1 is a betacoronavirus virulence factor (Zhang et al., 2015).
5. Screening antiviral drugs using reverse genetics
A recombinant coronavirus would efficiently express green fluorescent protein (GFP), which is commonly applied as a tool for high-throughput drug screens and neutralizing antibody assays. Feline coronavirus (FCoV) is one of the most significant coronaviruses. Tissue culture-adapted type I FCoV often loses pathogenicity, which complicates research on type I FCoV-induced feline infectious peritonitis (FIP). Therefore, researchers established a recombinant reporter C3663 virus carrying the nanoluciferase (Nluc) gene. They examined the inhibitory effect of 68 compounds on C3663 replication in Fcwf-4 cells and infectivity in a canine-derived cell line by using the reporter C3663 virus. Finally, they successfully screened a canine cell line, A72, that permitted FCoV replication but with low efficiency and aberrant viral gene expression (Terada et al., 2019).
6. Conclusion and future directions
Coronaviruses have repeatedly crossed species barriers, and some have emerged as important human pathogens. The best-known examples include SARS-CoV, MERS-CoV and SARS-CoV-2 (Chan et al., 2020), all of which pose a marked threat to human health. In addition, four other coronaviruses are known to infect humans, including HCoV-229E, HKU-NL63, HCoV-OC43 and HCoV-HKU1, which cause mild illnesses (Li et al., 2020a; Yan et al., 2020). Bats are generally thought to be the natural reservoir of a range of coronaviruses (Graham et al., 2013). Some bat coronaviruses also have the potential to emerge in human populations (Letko et al., 2020), and these include the SARS-like virus SHC014-CoV in horseshoe bats (Menachery et al., 2015). Human coronaviruses likely originated from bats and then jumped into another amplification mammalian host before crossing species barriers to infect humans. In addition to coronaviruses harming human health, the new emergence of animal coronaviruses has been a threat to animal health in the past decade. For example, porcine enteric alphacoronavirus, PEDV and SADS-CoV (Tan et al., 2020; Zaki et al., 2012; Zhou et al., 2018). To date, there are no clinically approved vaccines or antiviral drugs available for these coronavirus infections. An ideal live attenuated vaccine should replicate effectively in the host, not causing diseases, but stimulating enough protective immune responses without reverting to a virulent phenotype.
One of the most promising applications of reverse genetics is to realize the development of live attenuated vaccines by mutating or deleting virulence-related genes. To produce the ideal coronavirus vaccine strain through reverse genetics, a deeper knowledge of the coronavirus-host interaction, host immune responses, and pathogen immune evasion strategies is needed. For example, host cell pathways affected during coronavirus infection need to be identified, and these proteins of coronavirus suppressing the host innate immunity and the specific signaling pathway also need to be identified. Combining reverse genetics with metagenomics and structural biology can help characterize pre-emergent coronavirus populations, allowing the field to make predictions about which zoonotic coronaviruses are likely to emerge, prepare for future outbreaks, and facilitate the development of therapeutic strategies against coronavirus infection (Johnson et al., 2018). In addition, effective broad-spectrum antiviral drugs are still needed. Hopefully, live attenuated vaccines based on reverse genetics will be available in the near future and will prove to be highly effective tools against coronaviruses.
Acknowledgments
Support for this work was received from the National Key R&D Project of China (2017YFD0501102, 2017YFD0500605), Key R&D Project in Shaanxi Province of China (2019NY-076) and The Youth Innovation Team of Shaanxi Universities.
References
- Almazan F., Galan C., Enjuanes L. Engineering infectious cDNAs of coronavirus as bacterial artificial chromosomes. Methods Mol. Biol. 2008;454:275. doi: 10.1007/978-1-59745-181-9_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almazan F., DeDiego M.L., Sola I., Zuniga S., Nieto-Torres J.L., Marquez-Jurado S., Andres G., Enjuanes L. Engineering a replication-competent, propagation-defective Middle East respiratory syndrome coronavirus as a vaccine candidate. mBio. 2013;4 doi: 10.1128/mBio.00650-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almazan F., Gonzalez J.M., Penzes Z., Izeta A., Calvo r.E., Plana-Duran J., Enjuanes L. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. U. S. A. 2000;97:5516. doi: 10.1073/pnas.97.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelini M.M., Akhlaghpour M., Neuman B.W., Buchmeier M.J. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio. 2013;4 doi: 10.1128/mBio.00524-13. e00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balint A., Farsang A., Zadori Z., Hornyak A., Dencso L., Almazan F., Enjuanes L., Belak S. Molecular characterization of feline infectious peritonitis virus strain DF-2 and studies of the role of ORF3abc in viral cell tropism. J. Virol. 2012;86:6258. doi: 10.1128/JVI.00189-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beachboard D.C., Anderson-Daniels J.M., Denison M.R. Mutations across murine hepatitis virus nsp4 alter virus fitness and membrane modifications. J. Virol. 2015;89:2080. doi: 10.1128/JVI.02776-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall A., Yount B., Lin C.M., Hou Y., Wang Q., Saif L., Baric R. Characterization of a pathogenic full-length cDNA clone and transmission model for porcine epidemic diarrhea virus strain PC22A. mBio. 2016;7 doi: 10.1128/mBio.01451-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becares M., Pascual-Iglesias A., Nogales A., Sola I., Enjuanes L., Zuniga S. Mutagenesis of coronavirus nsp14 reveals its potential role in modulation of the innate immune response. J. Virol. 2016;90:5399. doi: 10.1128/JVI.03259-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M.M., Graham R.L., Donaldson E.F., Rockx B., Sims A.C., Sheahan T., Pickles R.J., Corti D., Johnston R.E., Baric R.S. Synthetic recombinant bat Sars-like coronavirus is infectious in cultured cells and in mice. Proc. Natl. Acad. Sci. U. S. A. 2008;105:19944. doi: 10.1073/pnas.0808116105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickerton E., Dowgier G., Britton P. Recombinant infectious bronchitis viruses expressing heterologous S1 subunits: potential for a new generation of vaccines that replicate in Vero cells. J. Gen. Virol. 2018;99:1681. doi: 10.1099/jgv.0.001167. [DOI] [PubMed] [Google Scholar]
- Bickerton E., Maier H.J., Stevenson-Leggett P., Armesto M., Britton P. The S2 subunit of infectious bronchitis virus beaudette is a determinant of cellular tropism. J. Virol. 2018;92 doi: 10.1128/JVI.01044-18. e01044-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet M., Lugari A., Posthuma C.C., Zevenhoven J.C., Bernard S., Betzi S., Imbert I., Canard B., Guillemot J.C., Lecine P., Pfefferle S., Drosten C., Snijder E.J., Decroly E., Morelli X. Coronavirus Nsp10, a critical co-factor for activation of multiple replicative enzymes. J. Biol. Chem. 2014;289:25783. doi: 10.1074/jbc.M114.577353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukreyev A., Lamirande E.W., Buchholz U.J., Vogel L.N., Elkins W.R., St Claire M., Murphy B.R., Subbarao K., Collins P.L. Mucosal immunisation of African green monkeys (Cercopithecus aethiops) with an attenuated parainfluenza virus expressing the SARS coronavirus spike protein for the prevention of SARS. Lancet. 2004;363:2122. doi: 10.1016/S0140-6736(04)16501-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X. COVID-19: immunopathology and its implications for therapy. Nat. Rev. Immunol. 2020;20:269–270. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case J.B., Li Y., Elliott R., Lu X., Graepel K.W., Sexton N.R., Smith E.C., Weiss S.R., Denison M.R. Murine hepatitis virus nsp14 exoribonuclease activity is required for resistance to innate immunity. J. Virol. 2018;92 doi: 10.1128/JVI.01531-17. e01531-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castano-Rodriguez C., Honrubia J.M., Gutierrez-Alvarez J., DeDiego M.L., Nieto-Torres J.L., Jimenez-Guardeno J.M., Regla-Nava J.A., Fernandez-Delgado R., Verdia-Baguena C., Queralt-Martin M., Kochan G., Perlman S., Aguilella V.M., Sola I., Enjuanes L. Role of severe acute respiratory syndrome coronavirus viroporins E, 3a, and 8a in replication and pathogenesis. mBio. 2018;9 doi: 10.1128/mBio.02325-17. e02325-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Kok K.H., Zhu Z., Chu H., To K.K., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol. 2017;39:529. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fehr A.R., Vijay R., Mack M., Zhao J., Meyerholz D.K., Perlman S. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19:181. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Tian J., Li Z., Kang H., Zhang J., Huang J., Yin H., Hu X., Qu L. Feline infectious peritonitis virus Nsp5 inhibits type I interferon production by cleaving NEMO at multiple sites. Viruses. 2019;12:43. doi: 10.3390/v12010043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Zhao Y., Xu G., Zhang K., Jia W., Sun Y., Zhao J., Xue J., Hu Y., Zhang G. The S2 subunit of QX-type infectious bronchitis coronavirus spike protein is an essential determinant of neurotropism. Viruses. 2019;11:972. doi: 10.3390/v11100972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockrell A.S., Beall A., Yount B., Baric R. Efficient reverse genetic systems for rapid genetic manipulation of emergent and preemergent infectious coronaviruses. Methods Mol. Biol. 2017;1602:59. doi: 10.1007/978-1-4939-6964-7_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comar C.E., Goldstein S.A., Li Y., Yount B., Baric R.S., Weiss S.R. Antagonism of dsRNA-induced innate immune pathways by NS4a and NS4b accessory proteins during MERS coronavirus infection. mBio. 2019;10 doi: 10.1128/mBio.00319-19. e00319-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottam E.M., Whelband M.C., Wileman T. Coronavirus NSP6 restricts autophagosome expansion. Autophagy. 2014;10:1426. doi: 10.4161/auto.29309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day M. Covid-19: Italy confirms 11 deaths as cases spread from north. BMJ. 2020;368:m757. doi: 10.1136/bmj.m757. [DOI] [PubMed] [Google Scholar]
- Deming D.J., Graham R.L., Denison M.R., Baric R.S. MHV-A59 ORF1a replicase protein nsp7-nsp10 processing in replication. Adv. Exp. Med. Biol. 2006;581:101. doi: 10.1007/978-0-387-33012-9_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., StJohn S.E., Osswald H.L., O'Brien A., Banach B.S., Sleeman K., Ghosh A.K., Mesecar A.D., Baker S.C. Coronaviruses resistant to a 3C-like protease inhibitor are attenuated for replication and pathogenesis, revealing a low genetic barrier but high fitness cost of resistance. J. Virol. 2014;88:11886. doi: 10.1128/JVI.01528-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Hackbart M., Mettelman R.C., O'Brien A., Mielech A.M., Yi G., Kao C.C., Baker S.C. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc. Natl. Acad. Sci. U. S. A. 2017;114 doi: 10.1073/pnas.1618310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., van Geelen A., Buckley A.C., O'Brien A., Pillatzki A., Lager K.M., Faaberg K.S., Baker S.C. Coronavirus endoribonuclease activity in porcine epidemic diarrhea virus suppresses type I and type III interferon responses. J. Virol. 2019;93:e02000-18. doi: 10.1128/JVI.02000-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Fang L., Yuan S., Zhao L., Wang X., Long S., Wang M., Wang D., Foda M.F., Xiao S. The nucleocapsid proteins of mouse hepatitis virus and severe acute respiratory syndrome coronavirus share the same IFN-β antagonizing mechanism: attenuation of PACT-mediated RIG-I/MDA5 activation. Oncotarget. 2017;8:49655. doi: 10.18632/oncotarget.17912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Dutta N.K., Mazumdar K., Lee B.H., Baek M.W., Kim D.J., Na Y.R., Park S.H., Lee H.K., Kariwa H., Le Q.M. Search for potential target site of nucleocapsid gene for the design of an epitope-based SARS DNA vaccine. Immunol. Lett. 2008;118:65. doi: 10.1016/j.imlet.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff M.P., Ferron F., Campanacci V., Longhi S., Rancurel C., Dutartre H., Snijder E.J., Gorbalenya A.E., Cambillau C., Canard B. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc. Natl. Acad. Sci. U. S. A. 2004;101:3792. doi: 10.1073/pnas.0307877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis S., Keep S., Britton P., de Wit S., Bickerton E., Vervelde L. Recombinant infectious bronchitis viruses expressing chimeric spike glycoproteins induce partial protective immunity against homologous challenge despite limited replication in vivo. J. Virol. 2018;92 doi: 10.1128/JVI.01473-18. e01473-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P., Fang L., Ren J., Hong Y., Liu X., Zhao Y., Wang D., Peng G., Xiao S. Porcine deltacoronavirus accessory protein NS6 antagonizes interferon beta production by interfering with the binding of RIG-I/MDA5 to double-stranded RNA. J. Virol. 2018;92 doi: 10.1128/JVI.00712-18. e00712-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R. Bacterial artificial chromosome-based lambda red recombination with the I-SceI homing endonuclease for genetic alteration of MERS-CoV. Methods Mol. Biol. 2020;2099:53. doi: 10.1007/978-1-0716-0211-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Athmer J., Channappanavar R., Phillips J.M., Meyerholz D.K., Perlman S. The nsp3 macrodomain promotes virulence in mice with coronavirus-induced encephalitis. J. Virol. 2015;89:1523. doi: 10.1128/JVI.02596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Channappanavar R., Jankevicius G., Fett C., Zhao J., Athmer J., Meyerholz D.K., Ahel I., Perlman S. The conserved coronavirus macrodomain promotes virulence and suppresses the innate immune response during severe acute respiratory syndrome coronavirus infection. mBio. 2016;7 doi: 10.1128/mBio.01721-16. e01721-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fett C., DeDiego M.L., Regla-Nava J.A., Enjuanes L., Perlman S. Complete protection against severe acute respiratory syndrome coronavirus-mediated lethal respiratory disease in aged mice by immunization with a mouse-adapted virus lacking E protein. J. Virol. 2013;87:6551. doi: 10.1128/JVI.00087-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graepel K.W., Lu X.T., Case J.B., Sexton N.R., Smith E.C., Denison M.R. Proofreading-deficient coronaviruses adapt for increased fitness over long-term passage without reversion of exoribonuclease-inactivating mutations. mBio. 2017;8:e01503-17. doi: 10.1128/mBio.01503-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R.L., Sims A.C., Brockway S.M., Baric R.S., Denison M.R. The nsp2 replicase proteins of murine hepatitis virus and severe acute respiratory syndrome coronavirus are dispensable for viral replication. J. Virol. 2005;79:13399. doi: 10.1128/JVI.79.21.13399-13411.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R.L., Becker M.M., Eckerle L.D., Bolles M., Denison M.R., Baric R.S. A live, impaired-fidelity coronavirus vaccine protects in an aged, immunocompromised mouse model of lethal disease. Nat. Med. 2012;18:1820. doi: 10.1038/nm.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R.L., Donaldson E.F., Baric R.S. A decade after SARS: strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 2013;11:836. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haijema B.J., Volders H., Rottier P.J. Switching species tropism: an effective way to manipulate the feline coronavirus genome. J. Virol. 2003;77:4528. doi: 10.1128/JVI.77.8.4528-4538.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Zhou Y., Siddiqui P., Niu J., Jiang S. Identification of immunodominant epitopes on the membrane protein of the severe acute respiratory syndrome-associated coronavirus. J. Clin. Microbiol. 2005;43:3718. doi: 10.1128/JCM.43.8.3718-3726.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotez P.J., Bottazzi M.E., Corry D.B. The potential role of Th17 immune responses in coronavirus immunopathology and vaccine-induced immune enhancement. Microbes Infect. 2020;22:165–167. doi: 10.1016/j.micinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Lin C.M., Yokoyama M., Yount B.L., Marthaler D., Douglas A.L., Ghimire S., Qin Y., Baric R.S., Saif L.J., Wang Q. Deletion of a 197-amino-acid region in the N-terminal domain of spike protein attenuates porcine epidemic diarrhea virus in piglets. J. Virol. 2017;91:e00227-17. doi: 10.1128/JVI.00227-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Ke H., Kim J., Yoo D., Su Y., Boley P., Chepngeno J., Vlasova A.N., Saif L.J., Wang Q. Engineering a live attenuated porcine epidemic diarrhea virus vaccine candidate via inactivation of the viral 2'-O-methyltransferase and the endocytosis signal of the spike protein. J. Virol. 2019;93:e00406-19. doi: 10.1128/JVI.00406-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Meulia T., Gao X., Saif L.J., Wang Q. Deletion of both the tyrosine-based endocytosis signal and the endoplasmic reticulum retrieval signal in the cytoplasmic tail of spike protein attenuates porcine epidemic diarrhea virus in pigs. J. Virol. 2019;93:e01758-18. doi: 10.1128/JVI.01758-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Li W., Gao T., Cui Y., Jin Y., Li P., Ma Q., Liu X., Cao C., Perlman S. The severe acute respiratory syndrome coronavirus nucleocapsid inhibits type I interferon production by interfering with TRIM25-mediated RIG-I ubiquitination. J. Virol. 2017;91 doi: 10.1128/JVI.02143-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernigan D.B., Team C.C.-R. Update: public health response to the coronavirus disease 2019 outbreak—United States, February 24, 2020. Morb. Mortal. Wkly Rep. 2020;69:216. doi: 10.15585/mmwr.mm6908e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W., Wang W., Zhao X., Zai J., Li X. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J. Med. Virol. 2020;92:433. doi: 10.1002/jmv.25682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Guardeno J.M., Nieto-Torres J.L., DeDiego M.L., Regla-Nava J.A., Fernandez-Delgado R., Castano-Rodriguez C., Enjuanes L. The PDZ-binding motif of severe acute respiratory syndrome coronavirus envelope protein is a determinant of viral pathogenesis. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Guardeno J.M., Regla-Nava J.A., Nieto-Torres J.L., DeDiego M.L., Castano-Rodriguez C., Fernandez-Delgado R., Perlman S., Enjuanes L. Identification of the mechanisms causing reversion to virulence in an attenuated SARS-CoV for the design of a genetically stable vaccine. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S., Kim S., Shin D.H., Kim M.S. Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzyme Inhib. Med. Chem. 2020;35:145. doi: 10.1080/14756366.2019.1690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B.A., Graham R.L., Menachery V.D. Viral metagenomics, protein structure, and reverse genetics: key strategies for investigating coronaviruses. Virology. 2018;517:30. doi: 10.1016/j.virol.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadkhoda K. COVID-19: an immunopathological view. mSphere. 2020;5:e00344-20. doi: 10.1128/mSphere.00344-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao C.F., Chang H.W. Investigation of the role of the spike protein in reversing the virulence of the highly virulent Taiwan porcine epidemic diarrhea virus Pintung 52 strains and its attenuated counterpart. Viruses. 2019;12:41. doi: 10.3390/v12010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Takayama-Ito M., Iizuka-Shiota I., Fukushi S., Posadas-Herrera G., Horiya M., Satoh M., Yoshikawa T., Yamada S., Harada S., Fujii H., Shibamura M., Inagaki T., Morimoto K., Saijo M., Lim C.K. Development of a recombinant replication-deficient rabies virus-based bivalent-vaccine against MERS-CoV and rabies virus and its humoral immunogenicity in mice. PLoS One. 2019;14 doi: 10.1371/journal.pone.0223684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.H., Kim H.J., Chang J. Superior immune responses induced by intranasal immunization with recombinant adenovirus-based vaccine expressing full-length spike protein of Middle East respiratory syndrome coronavirus. PLoS One. 2019;14 doi: 10.1371/journal.pone.0220196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kint J., Langereis M.A., Maier H.J., Britton P., van Kuppeveld F.J., Koumans J., Wiegertjes G.F., Forlenza M. Infectious bronchitis coronavirus limits interferon production by inducing a host shutoff that requires accessory protein 5b. J. Virol. 2016;90:7519. doi: 10.1128/JVI.00627-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong N., Shan T., Wang H., Jiao Y., Zuo Y., Li L., Tong W., Yu L., Jiang Y., Zhou Y., Li G., Gao F., Yu H., Zheng H., Tong G. BST2 suppresses porcine epidemic diarrhea virus replication by targeting and degrading virus nucleocapsid protein with selective autophagy. Autophagy. 2019:1. doi: 10.1080/15548627.2019.1707487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky-Bromberg S.A., Martinez-Sobrido L., Frieman M., Baric R.A., Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 2007;81:548. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y., Bae S., Myoung J. Middle East respiratory syndrome coronavirus-encoded accessory proteins impair MDA5-and TBK1-mediated activation of NF-kappaB. J. Microbiol. Biotechnol. 2019;29:1316. doi: 10.4014/jmb.1908.08004. [DOI] [PubMed] [Google Scholar]
- Lei L., Ying S., Baojun L., Yi Y., Xiang H., Wenli S., Zounan S., Deyin G., Qingyu Z., Jingmei L., Guohui C. Attenuation of mouse hepatitis virus by deletion of the LLRKxGxKG region of Nsp1. PLoS One. 2013;8 doi: 10.1371/journal.pone.0061166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J., Kusov Y., Hilgenfeld R. Nsp3 of coronaviruses: structures and functions of a large multi-domain protein. Antiviral Res. 2018;149:58. doi: 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Li H., Liu Y., Pan Y., Deng F., Song Y., Tang X., He Q. New variants of porcine epidemic diarrhea virus, China, 2011. Emerg. Infect. Dis. 2012;18:1350. doi: 10.3201/eid1808.120002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Li Z., Zou Y., Wicht O., van Kuppeveld F.J., Rottier P.J., Bosch B.J. Manipulation of the porcine epidemic diarrhea virus genome using targeted RNA recombination. PLoS One. 2013;8 doi: 10.1371/journal.pone.0069997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Jin Z., Gao Y., Zhou L., Ge X., Guo X., Han J., Yang H. Development of the full-length cDNA clones of two porcine epidemic diarrhea disease virus isolates with different virulence. PLoS One. 2017;12 doi: 10.1371/journal.pone.0173998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.Y., You Z., Wang Q., Zhou Z.J., Qiu Y., Luo R., Ge X.Y. The epidemic of 2019-novel-coronavirus (2019-nCoV) pneumonia and insights for emerging infectious diseases in the future. Microbes Infect. 2020;22:80–85. doi: 10.1016/j.micinf.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Li Z., Wohlford-Lenane C., Meyerholz D.K., Channappanavar R., An D., Perlman S., McCray P.B., Jr., He B. Single-dose, intranasal immunization with recombinant parainfluenza virus 5 expressing Middle East respiratory syndrome coronavirus (MERS-CoV) spike protein protects mice from fatal MERS-CoV infection. mBio. 2020;11:e00554-20. doi: 10.1128/mBio.00554-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokugamage K.G., Narayanan K., Nakagawa K., Terasaki K., Ramirez S.I., Tseng C.T., Makino S. Middle East respiratory syndrome coronavirus nsp1 inhibits host gene expression by selectively targeting mRNAs transcribed in the nucleus while sparing mRNAs of cytoplasmic origin. J. Virol. 2015;89:10970. doi: 10.1128/JVI.01352-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Pan J.A., Tao J., Guo D. SARS-CoV nucleocapsid protein antagonizes IFN-β response by targeting initial step of IFN-β induction pathway, and its C-terminal region is critical for the antagonism. Virus Genes. 2011;42:37. doi: 10.1007/s11262-010-0544-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui P.Y., Wong L.Y.R., Fung C.L., Siu K.L., Yeung M.L., Yuen K.S., Chan C.P., Woo P.C.Y., Yuen K.Y., Jin D.Y. Middle East respiratory syndrome coronavirus M protein suppresses type I interferon expression through the inhibition of TBK1-dependent phosphorylation of IRF3. Emerg. Microbes Infect. 2016;5:e39. doi: 10.1038/emi.2016.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo F., Liao F.L., Wang H., Tang H.B., Yang Z.Q., Hou W. Evaluation of antibody-dependent enhancement of SARS-CoV infection in rhesus macaques immunized with an inactivated SARS-CoV vaccine. Virol. Sin. 2018;33:201. doi: 10.1007/s12250-018-0009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S., Rottier P.J. Coronavirus reverse genetics by targeted RNA recombination. Curr. Top. Microbiol. Immunol. 2005;287:133. doi: 10.1007/3-540-26765-4_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcbride R., Van Zyl M., Fielding B. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Josset L., Gralinski L.E., Scobey T., Agnihothram S., Katze M.G., Baric R.S. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2'-O-methyltransferase activity. J. Virol. 2014;88:4251. doi: 10.1128/JVI.03571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Debbink K., Agnihothram S., Gralinski L.E., Plante J.A., Graham R.L., Scobey T., Ge X.Y., Donaldson E.F., Randell S.H., Lanzavecchia A., Marasco W.A., Shi Z.L., Baric R.S. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 2015;21:1508. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Gralinski L.E., Mitchell H.D., Dinnon K.H., 3rd, Leist S.R., Yount B.L., Jr., Graham R.L., McAnarney E.T., Stratton K.G., Cockrell A.S., Debbink K., Sims A.C., Waters K.M., Baric R.S. Middle East respiratory syndrome coronavirus nonstructural protein 16 is necessary for interferon resistance and viral pathogenesis. mSphere. 2017;2:e00346-17. doi: 10.1128/mSphere.00346-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Mitchell H.D., Cockrell A.S., Gralinski L.E., Yount B.L., Jr., Graham R.L., McAnarney E.T., Douglas M.G., Scobey T., Beall A., Dinnon K., 3rd, Kocher J.F., Hale A.E., Stratton K.G., Waters K.M., Baric R.S. MERS-CoV accessory ORFs play key role for infection and pathogenesis. mBio. 2017;8:e00665-17. doi: 10.1128/mBio.00665-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Gralinski L.E., Mitchell H.D., Dinnon K.H., 3rd, Leist S.R., Yount B.L., Jr., McAnarney E.T., Graham R.L., Waters K.M., Baric R.S. Combination attenuation offers strategy for live attenuated coronavirus vaccines. J. Virol. 2018;92:e00710-18. doi: 10.1128/JVI.00710-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielech A.M., Deng X., Chen Y., Kindler E., Wheeler D.L., Mesecar A.D., Thiel V., Perlman S., Baker S.C. Murine coronavirus ubiquitin-like domain is important for papain-like protease stability and viral pathogenesis. J. Virol. 2015;89:4907. doi: 10.1128/JVI.00338-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster V.J., Koopmans M., van Doremalen N., van Riel D., de Wit E. A novel coronavirus emerging in China—key questions for impact assessment. N. Engl. J. Med. 2020;382:692. doi: 10.1056/NEJMp2000929. [DOI] [PubMed] [Google Scholar]
- Muth D., Meyer B., Niemeyer D., Schroeder S., Osterrieder N., Muller M.A., Drosten C. Transgene expression in the genome of Middle East respiratory syndrome coronavirus based on a novel reverse genetics system utilizing red-mediated recombination cloning. J. Gen. Virol. 2017;98:2461. doi: 10.1099/jgv.0.000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanaga K., Yamanouchi K., Fujiwara K. Protective effect of monoclonal antibodies on lethal mouse hepatitis virus infection in mice. J. Virol. 1986;59:168. doi: 10.1128/jvi.59.1.168-171.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiura H., Linton N.M., Akhmetzhanov A.R. Initial cluster of novel coronavirus (2019-nCoV) infections in Wuhan, China is consistent with substantial human-to-human transmission. J. Clin. Med. 2020;9:488. doi: 10.3390/jcm9020488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortego J., Escors D., Laude H., Enjuanes L. Generation of a replication-competent, propagation-deficient virus vector based on the transmissible gastroenteritis coronavirus genome. J. Virol. 2002;76:11518. doi: 10.1128/JVI.76.22.11518-11529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeri N.C., Shrestha N., Rahman M.S., Zaki R., Tan Z., Bibi S., Baghbanzadeh M., Aghamohammadi N., Zhang W., Haque U. The SARS, MERS and novel coronavirus (COVID-19) epidemics, the newest and biggest global health threats: what lessons have we learned? Int. J. Epidemiol. 2020 doi: 10.1093/ije/dyaa033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Q., Fang L., Ding Z., Wang D., Peng G., Xiao S. Rapid manipulation of the porcine epidemic diarrhea virus genome by CRISPR/Cas9 technology. J. Virol. Methods. 2020;276:113772. doi: 10.1016/j.jviromet.2019.113772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai Y., Kawachi K., Terada Y., Omori H., Matsuura Y., Kamitani W. Two-amino acids change in the nsp4 of SARS coronavirus abolishes viral replication. Virology. 2017;510:165. doi: 10.1016/j.virol.2017.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez C.M., Pascual-Iglesias A., Sola I., Zuniga S., Enjuanes L. Minimum determinants of transmissible gastroenteritis virus enteric tropism are located in the N-terminus of spike protein. Pathogens. 2019;9:2. doi: 10.3390/pathogens9010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16:69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scobey T., Yount B.L., Sims A.C., Donaldson E.F., Agnihothram S.S., Menachery V.D., Graham R.L., Swanstrom J., Bove P.F., Kim J.D., Grego S., Randell S.H., Baric R.S. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. U. S. A. 2013;110:16157. doi: 10.1073/pnas.1311542110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Wan Y., Liu C., Yount B., Gully K., Yang Y., Auerbach A., Peng G., Baric R., Li F. Structure of mouse coronavirus spike protein complexed with receptor reveals mechanism for viral entry. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang W., Yang Y., Rao Y., Rao X. The outbreak of SARS-CoV-2 pneumonia calls for viral vaccines. NPJ Vaccines. 2020;5:18. doi: 10.1038/s41541-020-0170-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehata M.M., Kandeil A., Mostafa A., Mahmoud S.H., Gomaa M.R., El-Shesheny R., Webby R., Kayali G., Ali M.A. A recombinant influenza a/H1N1 carrying a short immunogenic peptide of MERS-CoV as bivalent vaccine in BALB/c mice. Pathogens. 2019;8:281. doi: 10.3390/pathogens8040281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Ye G., Deng F., Wang G., Cui M., Fang L., Xiao S., Fu Z.F., Peng G. Structural basis for the inhibition of host gene expression by porcine epidemic diarrhea virus nsp1. J. Virol. 2018;92:e01896-17. doi: 10.1128/JVI.01896-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Wang G., Yang Y., Shi J., Fang L., Li F., Xiao S., Fu Z.F., Peng G. A conserved region of nonstructural protein 1 from alphacoronaviruses inhibits host gene expression and is critical for viral virulence. J. Biol. Chem. 2019;294:13606–13618. doi: 10.1074/jbc.RA119.009713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokri S., Mahmoudvand S., Taherkhani R., Farshadpour F. Modulation of the immune response by Middle East respiratory syndrome coronavirus. J. Cell. Physiol. 2019;234:2143. doi: 10.1002/jcp.27155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu K.L., Kok K.H., Ng M.H.J., Poon V.K.M., Yuen K.Y., Zheng B.J., Jin D.Y. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3.TANK.TBK1/IKKepsilon complex. J. Biol. Chem. 2009;284:16202. doi: 10.1074/jbc.M109.008227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.C., Case J.B., Blanc H., Isakov O., Shomron N., Vignuzzi M., Denison M.R., Perlman S. Mutations in coronavirus nonstructural protein 10 decrease virus replication fidelity. J. Virol. 2015;89:6418. doi: 10.1128/JVI.00110-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson G.W., Hoang H., Schwartz K.J., Burrough E.R., Sun D., Madson D., Cooper V.L., Pillatzki A., Gauger P., Schmitt B.J., Koster L.G., Killian M.L., Yoon K.J. Emergence of porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J. Vet. Diagn. Invest. 2013;25:649. doi: 10.1177/1040638713501675. [DOI] [PubMed] [Google Scholar]
- Stobart C.C., Moore M.L. RNA virus reverse genetics and vaccine design. Viruses. 2014;6:2531. doi: 10.3390/v6072531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Li Y., He J., Hu Y., Cai X., Liu W., Liu T., Wang J., Li Z., Yuan X., Zhan Y., Yang L., Deng Z., Wang N., Yang Y., Wang A. Epidemic and genetic characterization of porcine epidemic diarrhea virus strains circulating in the regions around Hunan, China, during 2017-2018. Arch. Virol. 2020;165:877–889. doi: 10.1007/s00705-020-04532-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada Y., Kuroda Y., Morikawa S., Matsuura Y., Maeda K., Kamitani W. Establishment of a virulent full-length cDNA clone for type I feline coronavirus strain C3663. J. Virol. 2019;93:e01208-19. doi: 10.1128/JVI.01208-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanh Le T., Andreadakis Z., Kumar A., Gomez Roman R., Tollefsen S., Saville M., Mayhew S. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19:305–306. doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- Thao T.T.N., Labroussaa F., Ebert N., V'Kovski P., Stalder H., Portmann J., Kelly J., Steiner S., Holwerda M., Kratzel A., Gultom M., Schmied K., Laloli L., Husser L., Wider M., Pfaender S., Hirt D., Cippa V., Crespo-Pomar S., Schroder S., Muth D., Niemeyer D., Corman V., Muller M.A., Drosten C., Dijkman R., Jores J., Thiel V. Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. Nature. 2020 doi: 10.1038/s41586-020-2294-9. [DOI] [PubMed] [Google Scholar]
- Thiel V., Siddell S.G. Reverse genetics of coronaviruses using vaccinia virus vectors. Curr. Top. Microbiol. Immunol. 2005;287:199. doi: 10.1007/3-540-26765-4_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Herold J., Schelle B., Siddell S.G. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J. Gen. Virol. 2001;82:1273. doi: 10.1099/0022-1317-82-6-1273. [DOI] [PubMed] [Google Scholar]
- Tomar S., Johnston M.L., St John S.E., Osswald H.L., Nyalapatla P.R., Paul L.N., Ghosh A.K., Denison M.R., Mesecar A.D. Ligand-induced dimerization of Middle East respiratory syndrome (MERS) coronavirus nsp5 protease (3CLpro): implications for nsp5 regulation and the development of antivirals. J. Biol. Chem. 2015;290:19403. doi: 10.1074/jbc.M115.651463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujike M., Huang C., Shirato K., Makino S., Taguchi F. The contribution of the cytoplasmic retrieval signal of severe acute respiratory syndrome coronavirus to intracellular accumulation of S proteins and incorporation of S protein into virus-like particles. J. Gen. Virol. 2016;97:1853. doi: 10.1099/jgv.0.000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Beurden S.J., Berends A.J., Krämer-Kühl A., Spekreijse D., Chenard G., Philipp H.C., Mundt E., Rottier P.J.M., Verheije M.H. Recombinant live attenuated avian coronavirus vaccines with deletions in the accessory genes 3ab and/or 5ab protect against infectious bronchitis in chickens. Vaccine. 2018;36:1085. doi: 10.1016/j.vaccine.2018.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Worm S.H., Eriksson K.K., Zevenhoven J.C., Weber F., Züst R., Kuri T., Dijkman R., Chang G., Siddell S.G., Snijder E.J., Thiel V. Reverse genetics of SARS-related coronavirus using Vaccinia virus-based recombination. PLoS One. 2012;7 doi: 10.1371/journal.pone.0032857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit S., Jany S., Fux R., Sutter G., Volz A. CD8 + T cells responding to the Middle East respiratory syndrome coronavirus nucleocapsid protein delivered by vaccinia virus MVA in mice. Viruses. 2018;10:718. doi: 10.3390/v10120718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Fang L., Shi Y., Zhang H., Gao L., Peng G., Chen H., Li K., Xiao S. Porcine epidemic diarrhea virus 3C-like protease regulates its interferon antagonism by cleaving NEMO. J. Virol. 2016;90:2090. doi: 10.1128/JVI.02514-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Zheng X., Gai W., Zhao Y., Wang H., Wang H., Feng N., Chi H., Qiu B., Li N., Wang T., Gao Y., Yang S., Xia X. MERS-CoV virus-like particles produced in insect cells induce specific humoural and cellular imminity in rhesus macaques. Oncotarget. 2017;8:12686. doi: 10.18632/oncotarget.8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Ge X., Chen D., Li J., Cai Y., Deng J., Zhou L., Guo X., Han J., Yang H. The S gene is necessary but not sufficient for the virulence of porcine epidemic diarrhea virus novel variant strain BJ2011C. J. Virol. 2018;92:e00603-18. doi: 10.1128/JVI.00603-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Qiao X., Zhang S., Qin Y., Guo T., Hao Z., Sun L., Wang X., Wang Y., Jiang Y., Tang L., Xu Y., Li Y. Porcine transmissible gastroenteritis virus nonstructural protein 2 contributes to inflammation via NF-kappaB activation. Virulence. 2018;9:1685. doi: 10.1080/21505594.2018.1536632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Liang R., Liu Z., Shen Z., Shi J., Shi Y., Deng F., Xiao S., Fu Z.F., Peng G. The N-terminal domain of spike protein is not the enteric tropism determinant for transmissible gastroenteritis virus in piglets. Viruses. 2019;11:313. doi: 10.3390/v11040313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Sun A., Sun Y., Zhang S., Xia T., Guo T., Hao Z., Sun L., Jiang Y., Qiao X., Cui W., Tang L., Xu Y., Li Y., Wang L. Porcine transmissible gastroenteritis virus inhibits NF-kappaB activity via nonstructural protein 3 to evade host immune system. Virol. J. 2019;16:97. doi: 10.1186/s12985-019-1206-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenaar T.M., Zou Y. 2019_nCoV/SARS-CoV-2: rapid classification of betacoronaviruses and identification of traditional Chinese medicine as potential origin of zoonotic coronaviruses. Lett. Appl. Microbiol. 2020;70:342–348. doi: 10.1111/lam.13285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H.H., Fung T.S., Fang S., Huang M., Le M.T., Liu D.X. Accessory proteins 8b and 8ab of severe acute respiratory syndrome coronavirus suppress the interferon signaling pathway by mediating ubiquitin-dependent rapid degradation of interferon regulatory factor 3. Virology. 2018;515:165. doi: 10.1016/j.virol.2017.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., Jiang T. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Muruato A., Lokugamage K.G., Narayanan K., Zhang X., Zou J., Liu J., Schindewolf C., Bopp N.E., Aguilar P.V., Plante K.S., Weaver S.C., Makino S., LeDuc J.W., Menachery V.D., Shi P.Y. An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe. 2020;27:841–848.e3. doi: 10.1016/j.chom.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L., Tai Y., Bai C., Gao T., Song J., Xia P., Dong J., Zhao J., Wang F.S. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Guan Z.Y., Zhu W.F., Zhong L.Y., Qiu Z.Q., Yue P.F., Wu W.T., Liu J., Huang X. Preparation of puerarin chitosan oral nanoparticles by ionic gelation method and its related kinetics. Pharmaceutics. 2020;12:216. doi: 10.3390/pharmaceutics12030216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong C.Y., Ong H.K., Yeap S.K., Ho K.L., Tan W.S. Recent advances in the vaccine development against Middle East respiratory syndrome-coronavirus. Front. Microbiol. 2019;10:1781. doi: 10.3389/fmicb.2019.01781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Curtis K.M., Baric R.S. Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model. J. Virol. 2000;74:10600. doi: 10.1128/jvi.74.22.10600-10611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Curtis K.M., Fritz E.A., Hensley L.E., Jahrling P.B., Prentice E., Denison M.R., Geisbert T.W., Baric R.S. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12995. doi: 10.1073/pnas.1735582100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B., Denison M.R., Weiss S.R., Baric R.S. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J Virol. 2002;76:11065. doi: 10.1128/JVI.76.21.11065-11078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y., Nabar N.R., Shi C.S., Kamenyeva O., Xiao X., Hwang I.Y., Wang M., Kehrl J.H. SARS-coronavirus open reading frame-3a drives multimodal necrotic cell death. Cell Death Dis. 2018;9:904. doi: 10.1038/s41419-018-0917-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zeng L.P., Gao Y.T., Ge X.Y., Zhang Q., Peng C., Yang X.L., Tan B., Chen J., Chmura A.A., Daszak P., Shi Z.L. Bat severe acute respiratory syndrome-like coronavirus WIV1 encodes an extra accessory protein, ORFX, involved in modulation of the host immune response. J. Virol. 2016;90:6573. doi: 10.1128/JVI.03079-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Li Y., Cowley T.J., Steinbrenner A.D., Phillips J.M., Yount B.L., Baric R.S., Weiss S.R. The nsp1, nsp13, and M proteins contribute to the hepatotropism of murine coronavirus JHM.WU. J. Virol. 2015;89:3598. doi: 10.1128/JVI.03535-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Shi K., Yoo D. Suppression of type I interferon production by porcine epidemic diarrhea virus and degradation of CREB-binding protein by nsp1. Virology. 2016;489:252. doi: 10.1016/j.virol.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Li L., Yan L., Ming Z., Jia Z., Lou Z., Rao Z. Structural and biochemical characterization of endoribonuclease Nsp15 encoded by Middle East respiratory syndrome coronavirus. J. Virol. 2018:92. doi: 10.1128/JVI.00893-18. e00893-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Ke H., Blikslager A., Fujita T., Yoo D. Type III interferon restriction by porcine epidemic diarrhea virus and the role of viral protein nsp1 in IRF1 signaling. J. Virol. 2018;92 doi: 10.1128/JVI.01677-17. e01677-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Li W., Zhou P., Liu D., Luo R., Jongkaewwattana A., He Q. Genetic manipulation of porcine deltacoronavirus reveals insights into NS6 and NS7 functions: a novel strategy for vaccine design. Emerg. Microbes Infect. 2020;9:20–31. doi: 10.1080/22221751.2019.1701391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Fan H., Lan T., Yang X.L., Shi W.F., Zhang W., Zhu Y., Zhang Y.W., Xie Q.M., Mani S., Zheng X.S., Li B., Li J.M., Guo H., Pei G.Q., An X.P., Chen J.W., Zhou L., Mai K.J., Wu Z.X., Li D., Anderson D.E., Zhang L.B., Li S.Y., Mi Z.Q., He T.T., Cong F., Guo P.J., Huang R., Luo Y., Liu X.L., Chen J., Huang Y., Sun Q., Zhang X.L., Wang Y.Y., Xing S.Z., Chen Y.S., Sun Y., Li J., Daszak P., Wang L.F., Shi Z.L., Tong Y.G., Ma J.Y. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556:255. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Wang D., Zhou J., Pan T., Chen J., Yang Y., Lv M., Ye X., Peng G., Fang L., Xiao S. Porcine deltacoronavirus nsp5 antagonizes type I interferon signaling by cleaving STAT2. J. Virol. 2017;91:e00003-17. doi: 10.1128/JVI.00003-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zust R., Cervantes-Barragan L., Kuri T., Blakqori G., Weber F., Ludewig B., Thiel V. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog. 2007;3:1062. doi: 10.1371/journal.ppat.0030109. [DOI] [PMC free article] [PubMed] [Google Scholar]