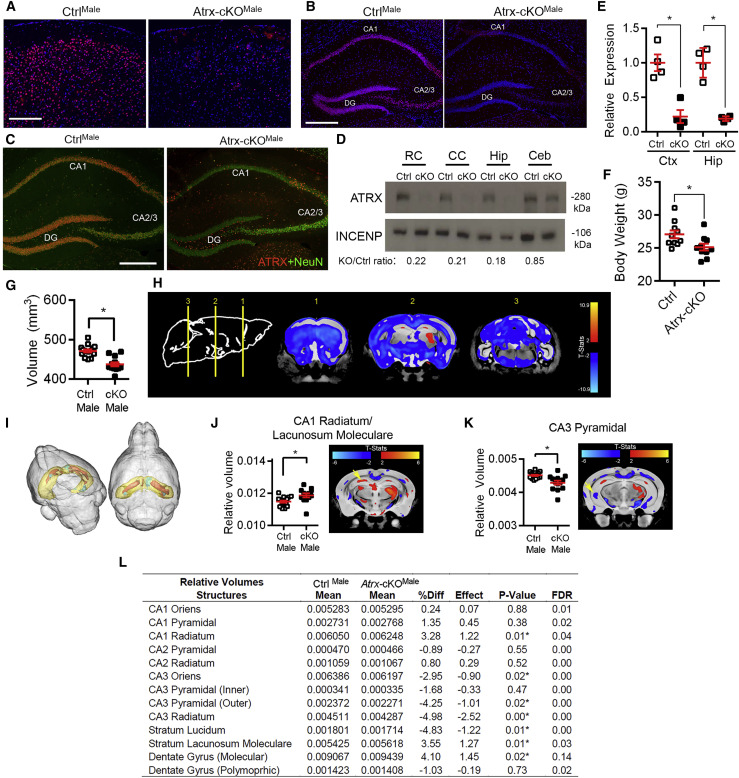
Figure 1.
Neuronal Inactivation of Atrx in Atrx-cKOMale Mice Causes Altered Hippocampal Morphology
(A and B) Immunofluorescence of the cortex (A) and hippocampus (B) of CtrlMale and Atrx-cKOMale mice. Scale bar: 100 μm for cortex, 200 μm for hippocampus.
(C) Co-staining of ATRX (red) and NeuN (green) shows absence of ATRX in hippocampal neurons in Atrx-cKOMale mice. CA1: cornu ammonis 1, CA2: cornu ammonis 2, CA3: cornu ammonis 3, DG: dentate gyrus. Scale bar: 200 μm.
(D) Western blot of ATRX in rostral cortex (RC), caudal cortex (CC), hippocampus (Hip), and cerebellum (Ceb) of CtrlMale and Atrx-cKOMale mice. The fold change in protein expression normalized to INCENP (loading control) is indicated below.
(E) Atrx RNA transcripts were measured by qRT-PCR in the rostral cortex (Ctx) (p < 0.005) and hippocampus (Hip) (p < 0.001) (n = 4 of each genotype).
(F) Body weight of CtrlMale and Atrx-cKOMale mice at 3 months of age (n = 10 each genotype; p < 0.05 by unpaired Student’s t test). MRI of 16 CtrlMale and 13 Atrx-cKOMale fixed mouse brains.
(G) Absolute total volume of CtrlMale and Atrx-cKOMale mouse brains (p < 0.0001).
(H) Coronal views of CtrlMale and Atrx-cKOMale brains show changes in the density of absolute volume.
(I) Cumulative 3-dimensional (3D) image generated from CtrlMale and Atrx-cKOMale mouse brains reveals altered hippocampal volume relative to brain size. Hippocampus is yellow, areas of increased volume in Atrx-cKOMale are orange, and areas of decreased volume are green.
(J) Relative volume of CA1 stratum radiatum and lacunosum moleculare between CtrlMale and Atrx-cKOMale mice (p < 0.05). MRI image at right shows cumulative changes in Atrx-cKOMale compared to CtrlMale mice.
(K) Relative size of CA3 pyramidal layer between CtrlMale and Atrx-cKOMale mice (p < 0.005). MRI image at right shows cumulative changes in Atrx-cKOMale mice compared to controls.
(L) Relative volumes of all hippocampal structures, including mean and standard deviation of CtrlMale and Atrx-cKOMale, percentage difference (%Diff), effect size, p value, and false discovery rate (FDR). Asterisks indicate p < 0.05.
See also Figure S1.