Figure 5.
Identification of Hippocampal Transcripts Differentially Expressed According to Sex and Genotype
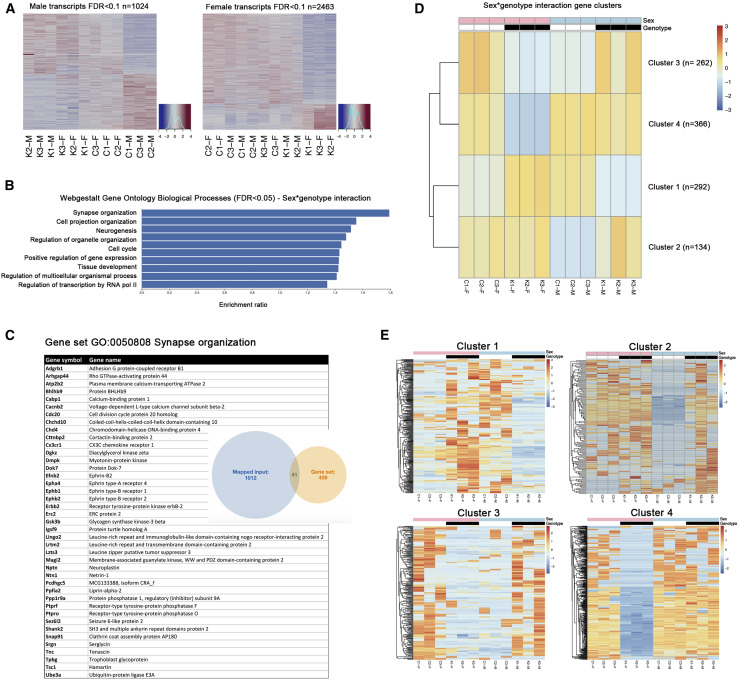
(A) RNA sequencing analysis was performed on CtrlMale(C-M), Atrx-cKOMale(K-M), CtrlFem(C-F), and Atrx-cKOFem(K-F) hippocampi (n = 3 each genotype), and an expression heatmap of the transcripts altered upon loss of ATRX was generated in male (left) and in female (right) mice, respectively (FDR < 0.1).
(B) Functional classification of sexually dimorphic transcripts identified in the hippocampus (biological processes, FDR < 0.05).
(C) List of genes included in the top category “synapse organization” identified by pathway analysis.
(D) Transcripts differentially expressed as a function of sex and genotype clustered into 4 categories. The number of genes in each cluster is indicated at right.
(E) Summary heatmaps of transcripts in clusters 1–4 identified in (D).