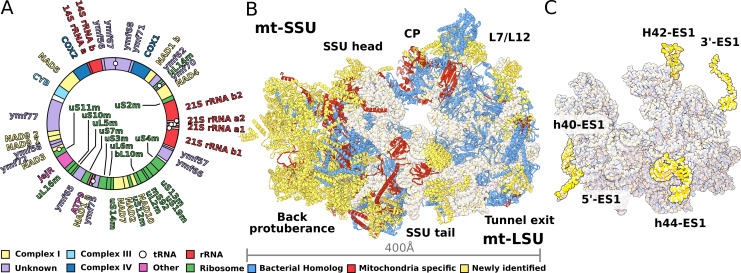
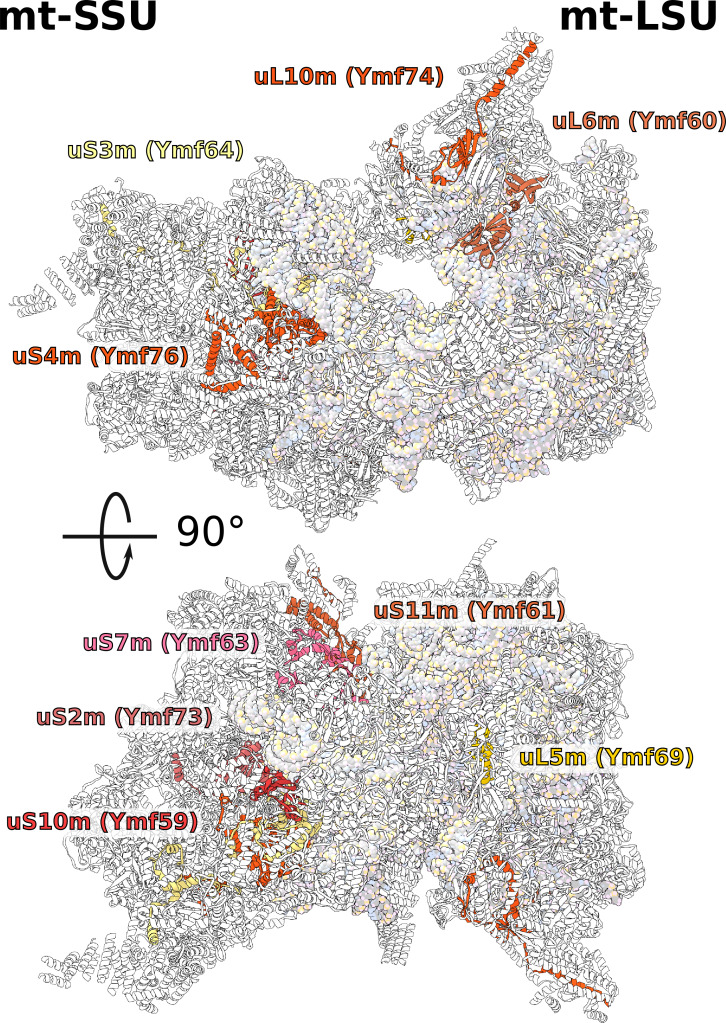
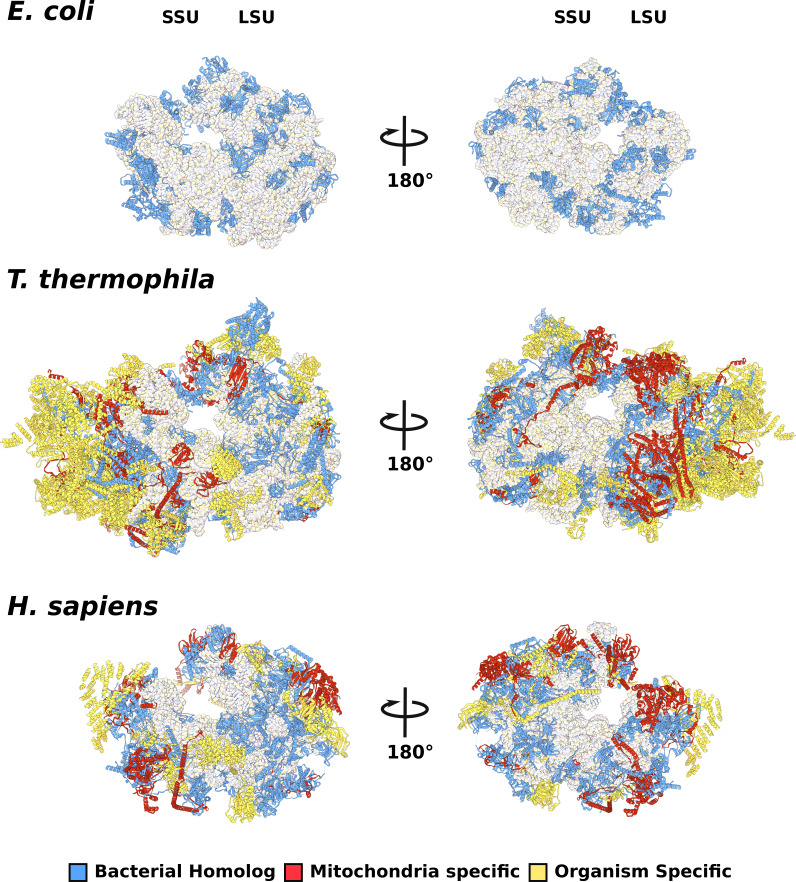
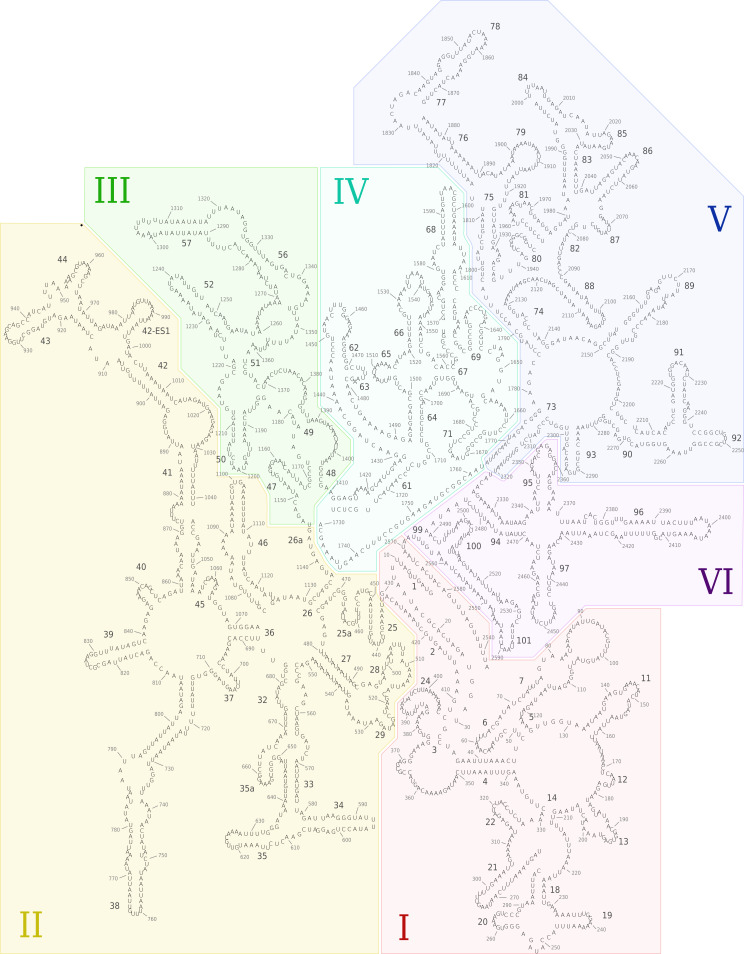
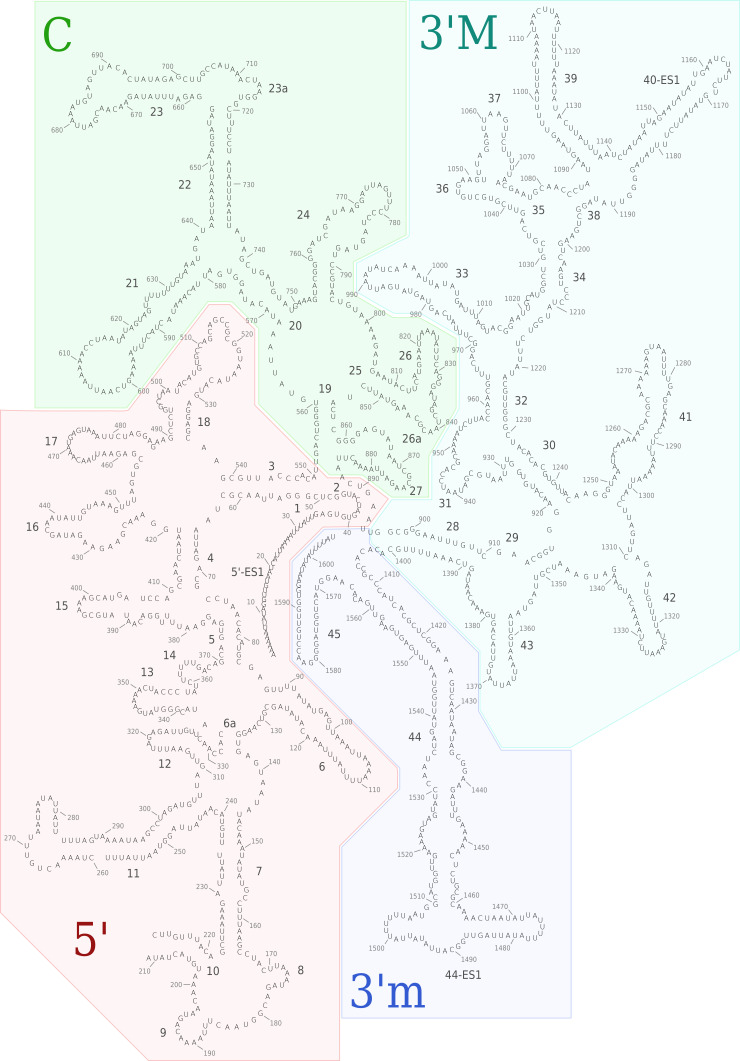
Figure 1. Structure of T. thermophila mitoribosome and newly identified proteins in the mitochondrial DNA.
(A) Schematic representation of the mitochondrial genome of T. thermophila with newly identified proteins labeled inside the circle. (B) The overall structure of the mitoribosome showing mitochondria specific and newly identified proteins. (C) Mitoribosomal rRNA showing expansion segments (relative to E. coli) in yellow.