Figure 3. Solid-phase extraction of tryptic peptides with triethylammonium acetate improves enrichment of ADP-ribosylated peptides.
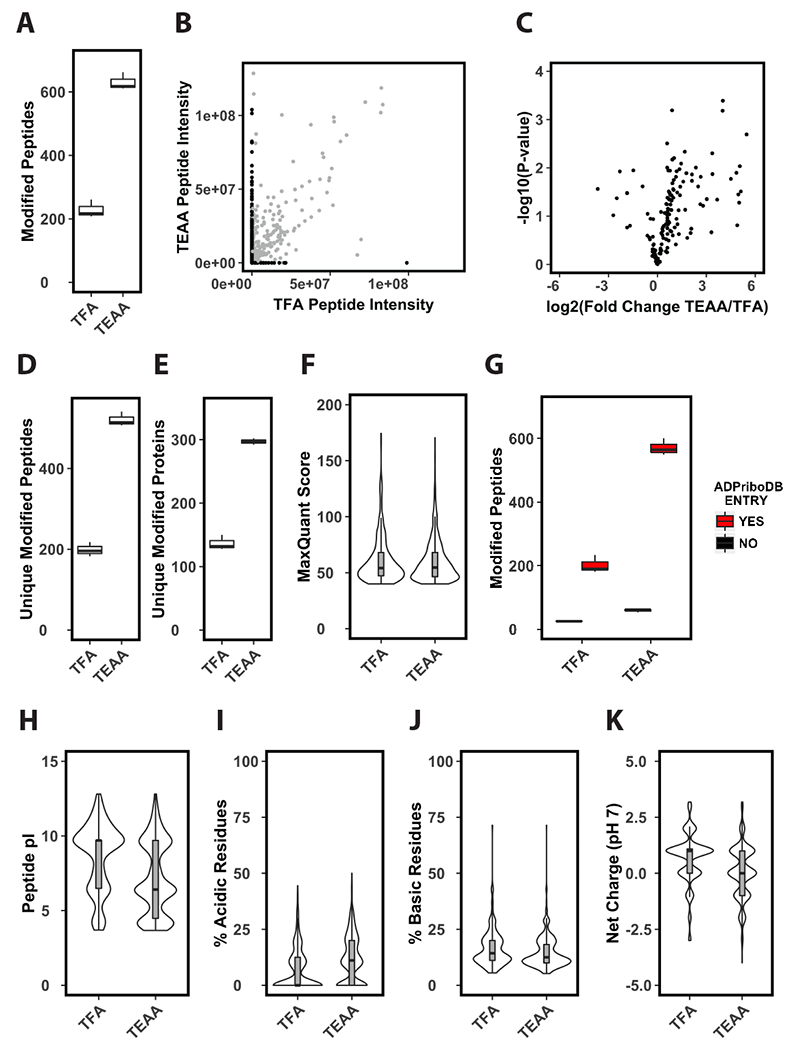
(A) Identifications per LC-MS/MS run of enriched phosphoribosylated peptides. Prior to enrichment tryptic cellular peptides were extracted by solid-phase using either TFA or TEAA as an ion-pairing reagent (n=3). (B) Average ion current intensity of enriched phosphoribosylated peptides. Peptides identified by both methods shown in grey, peptides identified by only one method are shown in black (n=3). (C) Volcano plot analysis of enriched phosphoribosylated peptides identified in both TFA and TEAA samples (n=3). (D-E) Unique identifications per LC-MS/MS run of enriched phosphoribosylated peptides (D) or proteins (E) (n=3). (F) Distribution of MaxQuant scores of phosphoribosylated peptides identified in TFA or TEAA extracted samples. (G) Identified phosphoribosylated peptides per LC-MS/MS runs in TFA or TEAA extracted samples that come from previously identified ADP-ribosylated substrates (n=3). This criterion is based on the presence (YES) or absence (NO) of the substrate from the curated ADP-ribosylated substrate database ADPriboDB. (H-K) Distribution of predicted pI values (H), percentage of acidic (I) or basic (J) residues, and average predicted net charge at pH 7 (K) of phosphoribosylated peptides identified in TFA or TEAA extracted samples.
