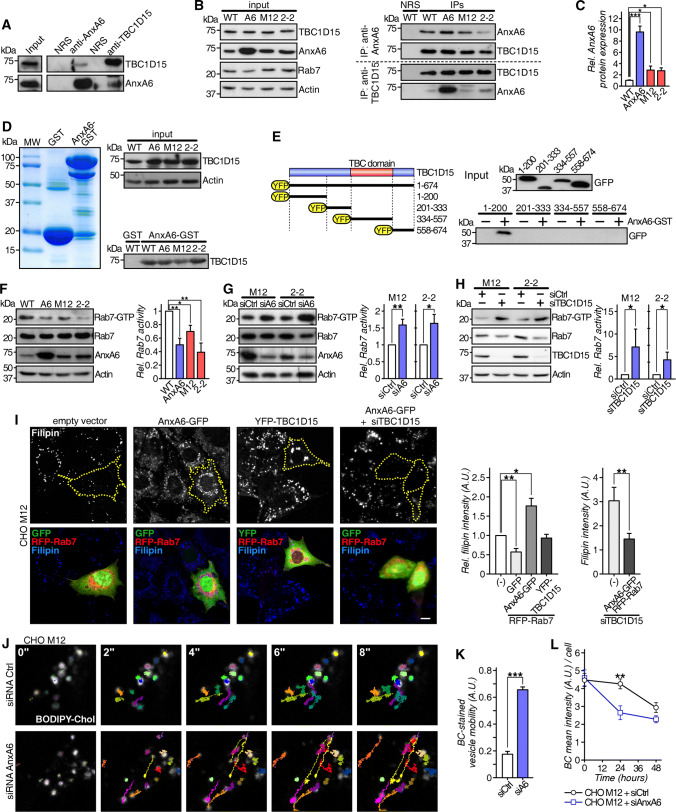
Fig. 2.
AnxA6 interacts with TBC1D15. a Mouse liver homogenates were immunoprecipitated with antibodies against AnxA6, TBC1D15 or control antibody (normal rabbit serum, NRS) and analyzed by western blotting for co-immunoprecipitation. Representative images of AnxA6 and TBC1D15 levels (5% of total input) and immunoprecipitations are shown (n = 2). b Cell lysates from CHO-WT, CHO-A6, CHO M12 and CHO 2-2 cells were immunoprecipitated with antibodies against AnxA6, TBC1D15 or control antibody (NRS) as indicated and analyzed by western blotting for co-immunoprecipitation. Representative images of TBC1D15, AnxA6, Rab7 and actin levels in the cell lysates (10% of total input, left panel) and immunoprecipitations are shown (n = 3). c Quantification of AnxA6 levels in CHO-WT, CHO-A6, CHO M12 and CHO 2-2 cell lysates (n = 3). d Coomassie blue staining of purified glutathione S-transferase (GST) and GST–AnxA6 used in the pull-down assays shown in D–E. Cell lysates (2% of total input) from CHO-WT, CHO-A6, CHO M12 and CHO 2-2 cells were incubated with GST or GST–AnxA6 fusion protein in pull-down assays and analyzed by western blotting with anti-TBC1D15 as indicated (n = 3). e Scheme of YFP-tagged TBC1D15 wild type (1–674aa) and deletion mutants (1–200, 201–333, 334–557, 558–674) used to map the interaction of TBC1D15 with AnxA6. The TBC domain in TBC1D15 (334–557) is indicated. Cell lysates from COS-1 cells ectopically expressing YFP–TBC1D15 wild type and mutants were incubated ± GST–AnxA6 fusion protein in pull-down assays and analyzed by western blotting for interaction with anti-GFP as indicated. Expression levels of YFP-tagged TBC1D15 deletion mutants (5% of total input) in COS-1 cell lysates are shown (n = 2). f Cell lysates from CHO-WT, CHO-A6, CHO M12 and CHO 2-2 cells were subjected to Rab interacting lysosomal protein (RILP)-C33–GST pull-down assays to determine active Rab7 (Rab7-GTP) levels. g Rab7-GTP levels determined as above with lysates from CHO M12 cells expressing non-targeting control siRNA (siCtrl) or siRNA targeting AnxA6 (siA6). Total levels of Rab7, AnxA6, and actin in cell lysates and the quantification of relative Rab7 activity are shown (5% of total input, n = 3). h Representative western blot showing Rab7-GTP levels determined as above with lysates from CHO M12 and CHO 2-2 cells expressing non-targeting control siRNA (siCtrl) or siRNA targeting TBC1D15 as indicated. Total levels of Rab7, TBC1D15 and actin in cell lysates (5% of total input) and the quantification of relative Rab7 activity are shown (n = 3). i CHO M12 cells were co-transfected with RFP–Rab7 (red) and empty vector (GFP), AnxA6–GFP, YFP–TBC1D15 or AnxA6–GFP together with siRNA targeting TBC1D15 (siTBC1D15) (green) as indicated. Cells were fixed and stained with filipin (blue). For better comparison of filipin staining, the outline and shape of transfected cells is indicated. Merged images are shown. Scale bar, 10 μm. The mean relative filipin intensity of at least 20 transfected cells from 3 independent experiments was quantified (n = 3). j Confocal images of CHO M12 cells expressing control siRNA (siRNA Ctrl) or siRNA targeting AnxA6 (siRNA AnxA6). Cells were grown in lipoprotein-protein deficient serum (LPDS), and then pulse-labeled with LDL-BODIPY-cholesteryl linoleate (see “Materials and methods” for details). Vesicles labeled with LDL-derived BODIPY-cholesterol (BC) were imaged in live cells over time. Representative images tracing pseudocoloured individual BC-stained vesicles for CHO M12 ± AnxA6 are shown (0–8 s) after 24 h chase. For representative frames from live-cell videos see Movies S1 and S2. k Quantitation of BC-stained vesicle mobility in CHO M12 ± AnxA6 (siCtr, siA6) after pulse-labeling with LDL-BODIPY-cholesteryl linoleate, followed by 24 h chase. Bars: 1 − [Pearson’s colocalization coefficient between subsequent frames/cell] ± SEM (n = 24–26 cells, two experiments). l Analysis of late endosomal (LE) BODIPY-cholesterol removal. CHO M12 cells were transfected with control siRNA (siCtrl) or siRNA targeting AnxA6 (siAnxA6). 22 h after transfection, cells were pulse-labeled for 2 h with 50 μg/ml LDL-BODIPY-cholesteryl linoleate. Cells were washed and chased in medium with 5% lipoprotein-deficient serum for 0–48 h. The efflux of LDL-derived BODIPY-cholesterol was quantified by analyzing mean fluorescence intensity per cell (n = 31–33 cells, two experiments). *p < 0.05; **p < 0.01; ***p < 0.001 by one-way ANOVA with Bonferroni post hoc test (c, f, i), two-tailed Student’s t test (g–i, k) or two-way ANOVA with Bonferroni post hoc test (l). All data are shown as mean ± SEM