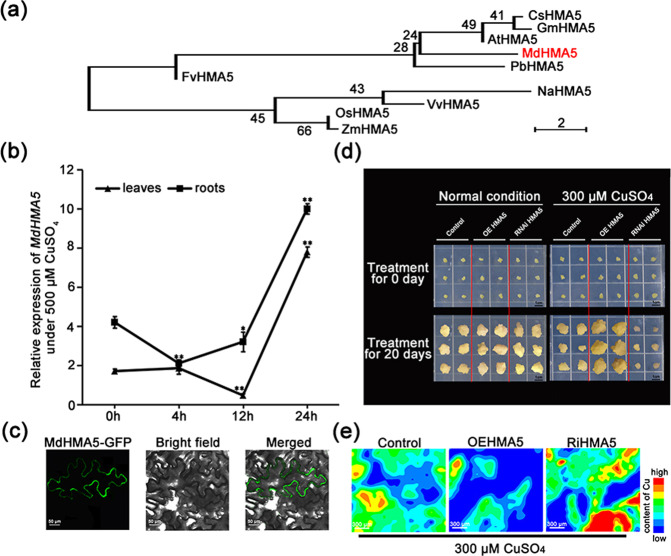
Fig. 5. Phylogenetic analysis, subcellular localization, and functional analysis of MdHMA5.
a Phylogenetic tree of the HMA5 subgroup of heavy metal ATPases from select plant species. The species and the accession numbers of the amino acid sequences used in the analysis are as follows: Arabidopsis thaliana (AT1G63440), Oryza sativa (OS04G46940), Fragaria vesca (FvH4_5g02091), Vitis vinifera (XP_010651259.1), Cucumis sativus (CSPI05G05530), Zea mays (ZM2G143512), Nicotiana attenuata (XP_019241377.1), Glycine max (GM11G024400), and Pyrus x bretschneideri (XP_018506630.1). bMdHMA5 expression in the leaves and roots under excess Cu treatment measured by qPCR and normalized according to the MdActin expression level. Hydroponic apple plants under excess Cu stress were sampled at 0, 4, 12, and 24h of treatment. The data are the means±SDs of triplicate experiments for each time point. The asterisks indicate the values that are significantly different from the controls (Student’s t-test): *P < 0.05; **P < 0.01. c MdHMA5-GFP is localized to the plasma membrane of Nicotiana benthamiana leaf epidermal cells. 35S::MdHMA5-GFP was transiently expressed in epidermal cells of N. benthamiana leaves and visualized by confocal microscopy (×40). d Cu tolerance of transgenic MdHMA5-overexpressing and MdHMA5-RNAi calli and untransformed control calli treated with excess Cu (300μM) for 20 days. e Cu levels, shown as μ-XRF elemental maps, in transgenic MdHMA5-overexpressing and MdHMA5-RNAi calli and control calli under excess Cu stress for 20 days