AOM by microorganisms limits the atmospheric release of the potent greenhouse gas methane and has consequent importance for the global carbon cycle and climate change modeling. While the oxidation of methane coupled to sulfate by consortia of anaerobic methanotrophic (ANME) archaea and bacteria is well documented, several other potential electron acceptors have also been reported to support AOM. In this study, we identify a number of novel respiratory strategies that appear to have been laterally acquired by members of the Methanoperedenaceae, as they are absent from related archaea and other ANME lineages. Expanding the known metabolic potential for members of the Methanoperedenaceae provides important insight into their ecology and suggests their role in linking methane oxidation to several global biogeochemical cycles.
KEYWORDS: ANME, AOM, comparative genomics, methane, Methanoperedenaceae
ABSTRACT
Anaerobic oxidation of methane (AOM) is an important biological process responsible for controlling the flux of methane into the atmosphere. Members of the archaeal family Methanoperedenaceae (formerly ANME-2d) have been demonstrated to couple AOM to the reduction of nitrate, iron, and manganese. Here, comparative genomic analysis of 16 Methanoperedenaceae metagenome-assembled genomes (MAGs), recovered from diverse environments, revealed novel respiratory strategies acquired through lateral gene transfer (LGT) events from diverse archaea and bacteria. Comprehensive phylogenetic analyses suggests that LGT has allowed members of the Methanoperedenaceae to acquire genes for the oxidation of hydrogen and formate and the reduction of arsenate, selenate, and elemental sulfur. Numerous membrane-bound multiheme c-type cytochrome complexes also appear to have been laterally acquired, which may be involved in the direct transfer of electrons to metal oxides, humic substances, and syntrophic partners.
INTRODUCTION
Anaerobic oxidation of methane (AOM) is an important microbiological process moderating the release of methane from anoxic waters and sediments into the atmosphere (1–4). Several diverse uncultured microbial lineages have been demonstrated to facilitate AOM. The bacterium “Candidatus Methylomirabilis oxyfera” is proposed to couple AOM to denitrification from nitrite, generating oxygen from nitric oxide for the activation of methane (5). Different lineages of anaerobic methanotrophic (ANME) archaea are hypothesized to mediate AOM through the reversal of the methanogenesis pathway and conserve energy using mechanisms similar to those found in methylotrophic and aceticlastic methanogens (6). Unlike methanogens, most of these ANMEs encode a large repertoire of multiheme c-type cytochromes (MHCs), which are proposed to mediate direct interspecies electron transfer to syntrophic sulfate-reducing bacteria (SRB) (7, 8) and/or the reduction of metal oxides and humic acids (9–12).
Currently, several clades within the archaeal phylum Euryarchaeota have been shown to be capable of anaerobic methanotrophy and include ANME-1a and -1b, ANME-2a to -2c, Methanoperedenaceae (formerly known as ANME-2d), and ANME-3 (13–15). Marine ANME lineages are often observed to form consortia with SRB, with ANME-1 and ANME-2 (a, b, and c) being associated with multiple genera within the Desulfobacterales and Desulfobulbaceae (13, 16–20), thermophilic ANME-1 being associated with “Candidatus Desulfofervidus auxilii” (8, 21), and ANME-3 being associated with SRBs of the Desulfobulbus (22). While members of the family Methanoperedenaceae have also recently been associated with SRB of the family Desulfobulbaceae in a freshwater lake sediment (23), they also appear to oxidize methane independently using a range of electron acceptors. The type species of this family, “Candidatus Methanoperedens nitroreducens,” was originally enriched in a bioreactor and shown to couple AOM to the reduction of nitrate via a laterally transferred nitrate reductase (15). Subsequently, “Candidatus Methanoperedens sp.” strain BLZ1 was also found to encode a laterally transferred nitrite reductase, which is also present in the genome of “Ca. Methanoperedens nitroreducens,” potentially allowing these microorganisms to couple AOM to dissimilatory nitrate reduction to ammonia (DNRA) (24). More recently, three novel species belonging to the Methanoperedenaceae were enriched in bioreactors demonstrated to couple AOM to the reduction of insoluble iron or manganese oxides (9, 12). These microorganisms did not encode dissimilatory nitrate reduction pathways but instead were inferred to use multiple unique MHCs during metal-dependent AOM to facilitate the transfer of electrons to the metal oxides (9, 12), consistent with the extracellular electron transfer mechanisms proposed for marine ANME organisms (7, 8). Bioreactor performance and 16S rRNA gene amplicon data have also been used to suggest that members of the Methanoperedenaceae are capable of AOM coupled to the reduction of selenate and chromium(VI), although this remains to be confirmed with more direct evidence (25, 26). Notably, members of the Methanoperedenaceae have been observed to facilitate AOM coupled to multiple terminal electron acceptors within the same natural sediment (27). Individual members of the family can possess such metabolic flexibility, with a lab-enriched species shown to couple AOM to the reduction of nitrate, iron, and manganese oxides (10). Given the relatively poor genomic representation of the Methanoperedenaceae and the lack of detailed physiological studies of its members, it is likely that considerable metabolic diversity for the lineage remains to be discovered.
In this study, comparative analysis was conducted on 16 Methanoperedenaceae metagenome-assembled genomes (MAGs) recovered from various environments to investigate the metabolic diversity and versatility of the family and to understand the evolutionary mechanisms responsible for these adaptations. These analyses indicate that members of the Methanoperedenaceae have acquired a large number of genes through lateral gene transfer (LGT) that potentially allow AOM to be coupled to a wide range of electron acceptors, suggesting that their role in methane oxidation extends beyond environments with nitrate and metal oxides.
RESULTS AND DISCUSSION
Expanding the genomic representation of the Methanoperedenaceae.
In order to explore the metabolic diversity within the Methanoperedenaceae, comparative genomic analysis was performed on both publicly available and newly acquired MAGs (Table 1). The publicly available genomes include six MAGs recovered from bioreactors where AOM is coupled to the reduction of nitrate (“Ca. Methanoperedens nitroreducens”; M.Nitro [15], BLZ2 [28], and IPS-1 [29]), iron (“Ca. Methanoperedens ferrireducens”; M.Ferri [9]), and manganese (“Ca. Methanoperedens manganicus” and “Ca. Methanoperedens manganireducens,” Mn-1 and Mn-2, respectively [12]). Also included are two environmental MAGs recovered from groundwater samples from the Horonobe and Mizunami underground research laboratories in Japan (HGW-1 and MGW-1) (30, 31). In order to recover additional genomes belonging to the family, GraftM (32) was used to screen public metagenome sequence data sets from the NCBI for Methanoperedenaceae-related 16S rRNA and mcrA gene sequences. Subsequent assembly and genome binning on data sets found to contain Methanoperedenaceae-like sequences led to the recovery of an additional eight MAGs belonging to the family. Six of these were from arsenic-contaminated groundwater samples (ASW-1-6), and a further two were from sediment and groundwater samples from a copper mine tailings dam (CMD-1 and CMD-2). All 16 MAGs are highly complete (≥87.4%), with low contamination (≤5.9%) based on 228 Euryarchaeota-specific marker genes (Table 1) (33). These genomes vary in GC content from 40.2 to 50.7% and range in size from 1.45 to 3.74 Mbp.
TABLE 1.
Characteristics of the metagenome-assembled genomes
| Bin ID | Genome size (mbp) |
No. of scaffolds |
N50
(scaffolds; bp) |
Strain hetero- geneitya |
Compl. (%)a |
Cont. (%)a |
%GC | No. of CDSse |
Source environment and associated publication |
Accession no.b | 16S rRNA gene? |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ASW-1 | 1.52 | 271 | 7,386 | 0.0 | 87.5 | 0.0 | 47.8 | 1,946 | Arsenic-contaminated groundwater, Bangladesh (104) |
SRR1563167, SRR1564103, SRR1573565, SRR1573578, SAMN10961276 |
N |
| ASW-2 | 2.63 | 157 | 28,058 | 25.0 | 94.4 | 4.8 | 48.0 | 2,944 | Arsenic-contaminated groundwater, Bangladesh (104) |
SRR1563167, SRR1564103, SRR1573565, SRR1573578, SAMN10961277 |
N |
| ASW-3 | 2.51 | 100 | 44,967 | 0.0 | 100.0 | 1.3 | 50.7 | 2,892 | Arsenic-contaminated groundwater, Bangladesh (104) |
SRR1563167, SRR1564103, SRR1573565, SRR1573578, SAMN10961278 |
N |
| ASW-4 | 2.24 | 155 | 24,336 | 0.0 | 97.1 | 0.7 | 43.2 | 2,464 | Arsenic-contaminated groundwater, Bangladesh (104) |
SRR1563167, SRR1564103, SRR1573565, SRR1573578, SAMN10961279 |
N |
| ASW-5 | 2.97 | 221 | 19,046 | 0.0 | 95.0 | 2.6 | 48.9 | 3,353 | Arsenic contaminated groundwater, Bangladesh (104) |
SRR1563167, SRR1564103, SRR1573565, SRR1573578, SAMN10961280 |
N |
| ASW-6 | 2.19 | 68 | 56,691 | 66.7 | 99.4 | 2.0 | 46.6 | 2,472 | Arsenic-contaminated groundwater, Bangladesh (104) |
SRR1563167, SRR1564103, SRR1573565, SRR1573578, SAMN10961281 |
Y |
| BLZ1c | 3.74 | 514 | 17,508 | 13.33 | 96.73 | 6.56 | 40.2 | 4,659 | AOM-nitrate bioreactor, Netherlands (24) |
LKCM00000000.1 | Y |
| BLZ2 | 3.74 | 85 | 74,304 | 0.0 | 99.4 | 4.6 | 40.3 | 4,041 | AOM-nitrate reactor, Netherlands (28) |
GCA_002487355.1 | N |
| CMD-1 | 1.85 | 116 | 27,949 | 100.0 | 98.0 | 0.7 | 44.9 | 2,261 | Copper mine tailings dam, Brazil (105) |
SRR5161805, SRR5161795, SAMN10961282 |
N |
| CMD-2 | 1.45 | 221 | 9,704 | 0.0 | 88.4 | 0.0 | 44.1 | 1,786 | Copper mine tailings dam, Brazil (105) |
SRR5161805, SRR5161795, SAMN10961283 |
N |
| HGW-1 | 2.00 | 128 | 24,496 | 33.3 | 96.4 | 2.0 | 43.2 | 2,288 | Groundwater samples, Japan (31) |
GCA_002839545.1 | Y |
| IPS-1 | 3.52 | 250 | 27,331 | 10.0 | 97.7 | 5.9 | 44.1 | 3,970 | AOM-nitrate bioreactor seeded from paddy field soil, Italy (29) |
GCA_900196725.1 | Y |
| M.Ferri | 2.91 | 59 | 88,069 | 0.0 | 98.7 | 1.3 | 40.8 | 3,019 | AOM-iron bioreactor, Australia (9) |
GCA_003104905.1 | Y |
| M.Nitro | 3.20 | 10 | 54,4976 | 0.0 | 99.7 | 1.3 | 43.2 | 3,428 | AOM-nitrate bioreactor, Australia (15) |
GCA_000685155.1 | Y |
| MGW-1 | 2.08 | 161 | 17,186 | 0.0 | 97.4 | 3.6 | 44.8 | 2,488 | Groundwater samples, Japan (30) |
Not availabled | N |
| Mn-1 | 3.59 | 68 | 87,551 | 0.0 | 100.0 | 1.3 | 40.6 | 3,737 | AOM-manganese bioreactor, Australia (12) |
SAMN10872768 | N |
| Mn-2 | 3.32 | 116 | 49,809 | 0.0 | 99.4 | 4.6 | 42.9 | 3,684 | AOM-manganese bioreactor, Australia (12) |
SAMN10872769 | N |
Completeness (compl.), contamination (cont.), and strain heterogeneity were estimated using CheckM (33).
Genome accession numbers. For the MAGs assembled in this study the SRA accession numbers are also given.
The BLZ1 genome was not used in analyses, as it is almost identical to the BLZ2 genome (99.5% ANI) and has inferior completeness and contamination values. The BLZ1 bioreactor was the parent system of the BLZ2 bioreactor.
This genome was provided by Yohey Suzuki and is associated with the study of Ino and colleagues (30).
CDSs, coding sequences; N, no; Y, yes.
A genome tree including 1,199 publicly available archaeal genomes, based on a concatenated set of 122 marker genes (34), confirmed the phylogenetic placement of the 16 MAGs within the Methanoperedenaceae. The genome tree supports that these MAGs form a monophyletic clade sister to the GoM-Arc1 genomes (Fig. 1). These genomes likely represent three separate genera within the family, based on their placement within a reference tree, relative evolutionary distance, FastANI distance, and average amino acid identity (AAI [35]; 61.3 to 89.2%) (see Fig. S1 in the supplemental material). All MAGs were classified as members of the genus “Ca. Methanoperedens,” except HGW-1 and ASW-3, which appear to represent independent genus-level lineages (Fig. 1). Phylogenetic analysis of the six MAGs containing 16S rRNA genes was consistent with the genome tree (Fig. S2), supporting their classification as members of the Methanoperedenaceae family.
FIG 1.
Phylogenetic placement of the Methanoperedenaceae MAGs and distribution of potential terminal electron acceptors. The genome tree was inferred using maximum likelihood with a concatenated set of 122 archaeon-specific marker genes. Black and white dots indicate >90% and >70% bootstrap values, respectively. The scale bar represents amino acid nucleotide changes. Based on GTDB-Tk, the family Methanoperedenaceae includes three genera, including “Ca. Methanoperedens,” which are denoted with brackets. The table to the right of the tree shows the presence/absence of genes associated with potential terminal electron acceptors in each corresponding Methanoperedenaceae genome.
Average amino acid identity (AAI%) for the Methanoperedenaceae genomes. AAI was calculated between each pair of genomes using CompareM. Download FIG S1, PDF file, 0.2 MB (172KB, pdf) .
Copyright © 2020 Leu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
16S rRNA gene-based phylogenetic placement of the Methanoperedenaceae MAGs. The 16S rRNA genes extracted from the Methanoperedenaceae MAGs from this study are indicated in red. Support values calculated via nonparametric bootstrapping. The scale bar represents numbers of changes per nucleotide position. Download FIG S2, PDF file, 0.5 MB (483.2KB, pdf) .
Copyright © 2020 Leu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Potential electron donors used by the Methanoperedenaceae.
Metabolic reconstruction of the Methanoperedenaceae MAGs showed that all genomes encoded the central methanogenesis pathway, inclusive of the methyl coenzyme M (methyl-CoM) reductase, supporting their potential for the complete oxidation of methane to CO2 (Fig. 2 and Fig. S3). The annotation of membrane-bound formate dehydrogenases (FdhAB) in four of the Methanoperedenaceae MAGs (Mn-2, ASW-4, ASW-1, and MGW-1) (Fig. 3) suggests that some members of the family may also oxidize formate (E0 [CO2/HCOO−] = −430 mV) (36). As the enzyme is reversible, these species may also potentially produce formate as a supplementary electron sink during AOM, as suggested for Mn-2 (12). Formate was proposed as a putative electron shuttle between ANME-1 organisms and their syntrophic partner SRB, based on the annotation and expression of fdhAB in ANME-1, but this has not been supported with physiological studies (37, 38). The putative formate dehydrogenase encoded in the Mn-2 MAG is phylogenetically related to an FdhA found in the genome of Caldiarchaeum subterraneum, while those encoded by ASW-4, ASW-1, and MGW-1 appear to be more similar to FdhA of Methanocellaceae archaeon UBA148 (Fig. 3).
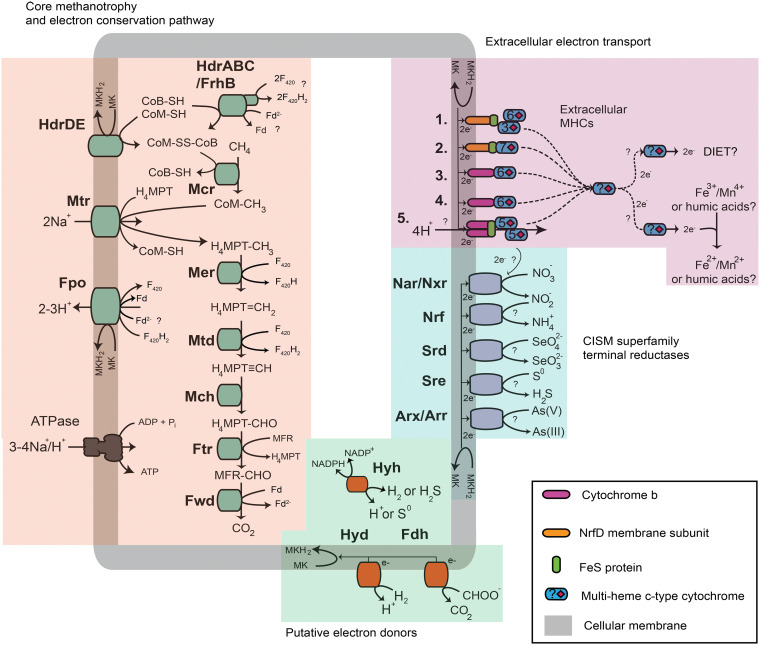
FIG 2.
Metabolic capabilities of the Methanoperedenaceae. Key metabolic pathways for the anaerobic oxidation of methane, energy conservation mechanisms, hydrogen and formate oxidation, and electron acceptors found within the pangenome of the Methanoperedenaceae. Numbers 1 to 5 indicate the different menaquinone:cytochrome c oxidoreductases conserved in the Methanoperedenaceae MAGs (Data Set S1A). Abbreviations for enzymes and cofactors in the figure are as follows: H4MPT, tetrahydromethanopterin; MFR, methanofuran; Fwd, formyl-methanofuran dehydrogenase; Ftr, formylmethanofuran/H4MPT formyltransferase; Mch, methenyl-H4MPT cyclohydrolase; Mtd, F420-dependent methylene H4MPT dehydrogenase; Mer, F420-dependent methylene-H4MPT reductase; Mtr, Na+-translocating methyl-H4MPT:coenzyme M (CoM) methyltransferase; Mcr, methyl-CoM reductase; F420, F420 coenzyme; Fd, ferredoxin; CoM-SH, coenzyme M; CoB-HS, coenzyme B; Hdr, heterodisulfide reductase; Fpo, F420H2 dehydrogenase; Hyd, type 1 NiFe hydrogenase; Hyh, type 3b NiFe hydrogenase; Fdh, formate dehydrogenase; Nar, nitrate reductase; Nrf, nitrite reductase; Srd, selenate reductase; Sre, sulfur reductase; Arx, arsenite oxidase; Arr, arsenate reductase; DIET, direct interspecies electron transfer.
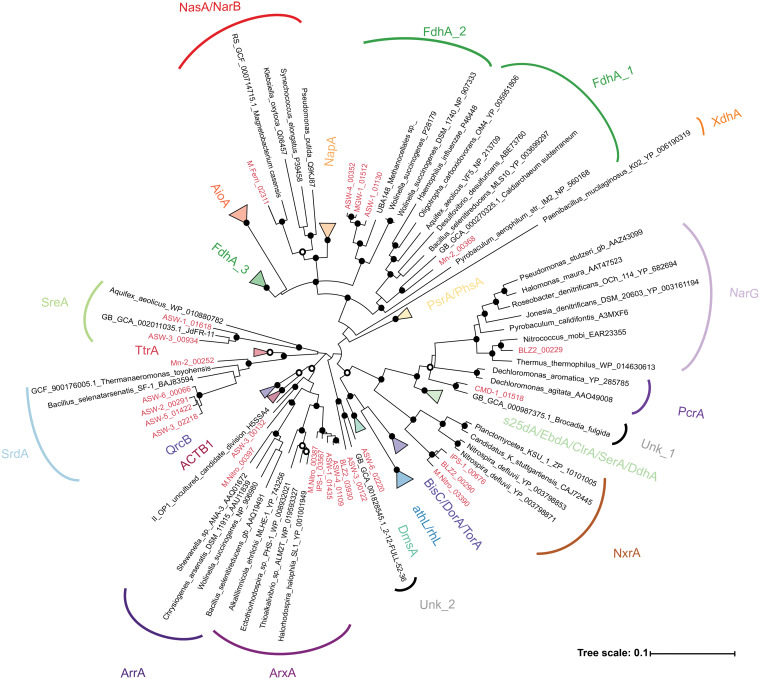
FIG 3.
Phylogenetic analysis of the catalytic subunits of the CISM superfamily. Putative genes recovered from the Methanoperedenaceae are indicated in red. The gene tree was inferred by maximum likelihood, and support values were calculated via nonparametric bootstrapping. Black and white dots indicate >90% and >70% bootstrap support, respectively. The scale bar represents amino acid changes. ACTB1, alternate complex III, domain of subunit B; ArrA, arsenate reductase; ArxA, arsenite oxidase; AthL, pyrogallol hydroxytransferase; BisC, biotin sulfoxide reductase; ClrA, chlorate reductase; EbdA, ethylbenzene dehydrogenase; s25dA, C25 dehydrogenase; DmsA, DMSO reductase; DorA, DMSO reductase; NapA, nitrate reductase; NarG, nitrate reductase; NasA, assimilatory nitrate reductase; NarB, assimilatory nitrate reductase; NxrA, nitrite oxidoreductase; PsrA, polysulfide reductase; PhsA, thiosulfate reductase; QrcB, quinone reductase complex; TtrA tetrathionate reductase; DmsA, PcrA, perchlorate reductase; SrdA, Selenate reductase; SreA, sulfur reductase; TorA, trimethylamine N-oxide (TMAO) reductase; XdhA, xanthine dehydrogenase; FdhA, formate dehydrogenase; rhL, resorcinol hydroxylase; Unk, unknown putative reductase. Amino acid sequences are included in Data Set S1B.
Phylogenetic analysis of methyl-coenzyme reductase subunit A (McrA). Putative genes recovered from the Methanoperedenaceae are indicated in red. The gene tree was inferred using maximum likelihood, and support values were calculated via nonparametric bootstrapping. The scale bar represents amino acid changes. Download FIG S3, PDF file, 0.1 MB (144.2KB, pdf) .
Copyright © 2020 Leu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sequences, identifiers, and statistics for genes used in the comparative analyses of the Methanoperedenaceae MAGs. (A) Genes encoding proteins involved in the methane oxidation pathway, energy conservation, and other metabolic pathways as shown in Fig. 2. (B) Amino acid sequences used in the CISM superfamily gene tree (Fig. 3). Amino acid sequences include curated sequences from Swiss-Prot and the work of Castelle et al. (C. J. Castelle, L. A. Hug, K. C. Wrighton, B. C. Thomas, et al., Nat Commun 4:2120, 2013, https://doi.org/10.1038/ncomms3120) and closely related sequences from GTDB r83 protein reference database (D. H. Parks, M. Chuvochina, D. W. Waite, C. Rinke, et al., Nat Biotechnol 36:996–1004, 2018, https://doi.org/10.1038/nbt.4229). (C) Amino acid sequences used in the catalytic subunits of the energy-converting NiFe hydrogenase. Amino acid sequences include curated sequences from Greening et al. (C. Greening, A. Biswas, C. R. Carere, C. J. Jackson, et al., ISME J 10:761–777, 2016, https://doi.org/10.1038/ismej.2015.153) and closely related sequences from the GTDB r83 protein reference database. (D) Genes encoding putative NiFe hydrogenase maturation proteins. (E) Best blastp hits of Fpo dehydrogenase subunits to the IMG database. Blastp hits show that divergent Fpo subunits are present in the Methanoperedenaceae MAGs, as seen in Fig. S6. Top blast hits to “Ca. Methanoperedens”-like protein sequences were excluded. (F) General statistic of multiheme c-type cytochromes (MHCs) in the ANME genomes. (G) MHC general statistics for all bacterial and archaeal families in the GTDB v89 database. Download Data Set S1, XLSX file, 0.2 MB (251.6KB, xlsx) .
Copyright © 2020 Leu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The use of hydrogen (H2; E0 = –414 mV [39]) as an electron source was previously suggested for MGW-1 and HGW-1, which encode group 1 membrane-bound NiFe hydrogenase complexes, composed of a NiFe catalytic subunit, a FeS electron transfer subunit, and a membrane-bound b-type cytochrome (30, 31). These hydrogenases, along with similar group 1 NiFe hydrogenases identified in the ASW-6 and CMD-2 MAGs, form a monophyletic clade with those encoded by the MAG for “Candidatus Hydrothermarchaeota” (JdFR-18), which belongs to the archaeal phylum Hydrothermarchaeota (40), and several members of the Halobacterota (Fig. S4A). The ASW-3 and ASW-5 MAGs encode group 1 NiFe hydrogenases that are basal to Vho/Vht/Vhx hydrogenases encoded by members of the genus Methanosarcina (41). As the ASW-5 NiFe hydrogenase does not encode a b-type cytochrome (Fig. S4B), it is unclear how electrons are derived from hydrogen. In addition to the membrane-bound NiFe hydrogenases, the M.Nitro MAG was found to carry genes for two different sets of group 3b cytoplasmic hydrogenases (Fig. S4A). The MGW-1 (30) and ASW-2 MAGs also encode group 3b hydrogenases, which have been implicated in hydrogen evolution and NADP (NADPH) reduction (42). Similar complexes have also been shown to have hydrogen oxidation and elemental-sulfur-reducing capabilities (42–44). It is unknown how these group 3b hydrogenases contribute to energy conservation given their predicted cytoplasmic localization. The functionality of the annotated group 1 and 3 NiFe hydrogenases is supported by the identification of the NiFe binding motifs (L1 and L2) on their NiFe catalytic subunits and the annotation of all or most of the hydrogenase maturation genes (hypA to -F) on the same Methanoperedenaceae MAGs (Data Set S1D). The potential for some Methanoperedenaceae to couple the oxidation of hydrogen and/or formate to the reduction of exogenous electron acceptors would be advantageous with the dynamic availability of methane in natural environments (45).
Phylogenetic analysis of the subunits of the NiFe hydrogenases annotated in the Methanoperedenaceae genomes. (A) Analysis of the catalytic subunits of the energy-converting NiFe hydrogenases. (B) Analysis of the b-type cytochrome in the group 1 NiFe hydrogenases. Putative genes recovered from the Methanoperedenaceae are indicated in red. The gene trees were inferred using maximum likelihood and support values calculated via nonparametric bootstrapping. The reference sequences of group 1 and group 3 NiFe hydrogenases were acquired from the work of Greening et al. (C. Greening, A. Biswas, C. R. Carere, C. J. Jackson, et al., ISME J 10:761–777, 2016, https://doi.org/10.1038/ismej.2015.153) and the GTDB v83 reference sequences (D. H. Parks, M. Chuvochina, D. W. Waite, C. Rinke, et al., Nat Biotechnol 36:996–1004, 2018, https://doi.org/10.1038/nbt.4229). The scale bars represent amino acid changes. Download FIG S4, PDF file, 0.7 MB (688KB, pdf) .
Copyright © 2020 Leu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Pathways for energy conservation during AOM in the Methanoperedenaceae.
All members of the Methanoperedenaceae encode the Fpo complex (FpoABCDHIJ1J2LMNOF), a homolog of complex I (nuoABCDEFGHIJKLMN), which is hypothesized to oxidize F420H2 coupled to the reduction of a membrane-bound soluble electron carrier and translocation of two protons out of the cell (Fig. 2 and Fig. S5A) (41, 46). While members of the Methanosarcinales and marine ANME-2a are reported to typically use methanophenazine (MP) as their membrane-bound soluble electron carrier, the Methanoperedenaceae and ANME-1 have previously been suggested to use menaquinone (MK) based on the annotation of the futalosine pathway for MK biosynthesis in several MAGs representing these lineages (47). Comparative genomic analysis of the 16 Methanoperedenaceae MAGs revealed that the futalosine pathway is a conserved feature of all members, except the most basal member, ASW-3 (see below and Data Set S1A). As has previously been suggested by Arshad et al. (24), the larger difference in redox potential between F420 (E0 = –360mV) and MK (E0 = –80mV [48]) than between F420 and MP (E0 = –165mV [49]) would theoretically allow the Fpo complex to translocate more protons (3H+/2e–) out of the cell for every molecule of F420 oxidized, giving a higher overall energetic yield from AOM (Fig. S5B).
Subunit compositions of the Fpo dehydrogenase protein complexes and theoretical bioenergetics of energy metabolism in ANME-2a and Methanoperedenaceae. (A) Fpo subunit components for the ANME-2a and ASW-3 genomes (top left) and the other members of the Methanoperedenaceae (bottom left). The utilization of different electron carriers shows greater biochemical energetic gains based on more potential proton translocation. The colors orange and green depict Methanosarcinales-like and non-Methanosarcinales-like subunits. (B) Theoretical redox potential drop when utilizing MP (left) or MK (right) during F420H2 and Fd2– oxidation. This is due to differences between the membrane-bound electron carriers’ redox midpoint potential (Em) of –80 mV and –165 mV for MK and MP, respectively (M. Tietze, A. Beuchle, I. Lamla, N. Orth, et al., Chembiochem 4:333–335, 2003, https://doi.org/10.1002/cbic.200390053; Q. H. Tran and G. Unden, Eur J Biochem 251:538–543, 1998, https://doi.org/10.1046/j.1432-1327.1998.2510538.x). Download FIG S5, PDF file, 0.3 MB (348.2KB, pdf) .
Copyright © 2020 Leu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phylogenetic analysis of the Fpo subunits annotated in the Methanoperedenaceae genomes. (A) FpoA; (B) FpoB; (C) FpoC; (D) FpoD; (E) FpoH; (F) FpoI; (G) FpoJ1; (H) FpoJ2; (I) FpoK; (J) FpoL; (K) FpoM; (L) FpoN; (M) FpoO. Putative genes recovered from the Methanoperedenaceae are indicated in red. The gene trees were inferred using maximum likelihood and support values calculated via nonparametric bootstrapping. Reference genes and the taxonomy are from the GTDB v83 database (D. H. Parks, M. Chuvochina, D. W. Waite, C. Rinke, et al., Nat Biotechnol 36:996–1004, 2018, https://doi.org/10.1038/nbt.4229). Download FIG S6, PDF file, 2.1 MB (2.1MB, pdf) .
Copyright © 2020 Leu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phylogenetic analysis of the Fpo complex in the Methanoperedenaceae MAGs showed that the FpoKLMNO subunits are homologous to proteins found in MP-utilizing members of the Methanosarcinales. The FpoABCDHIJ1J2 subunits are more similar to those found in microorganisms known to use MK and other quinones, which have more positive redox potentials (Fig. S5 and S6; Data Set S1E) (50). As the latter subunits (specifically FpoH) are responsible for the interaction with the membrane-soluble electron carrier pool (51, 52), this observation provides further support to the use of MK by members of the Methanoperedenaceae. To our knowledge, this is the first reported example of a lineage encoding a “hybrid” complex I homolog possessing subunits with homology to those found in phylogenetically diverse microorganisms (Fig. S6). The GoM-Arc-I MAGs appear to possess the MK biosynthesis pathway and a hybrid Fpo complex similar to those of the Methanoperedenaceae (Fig. S6), suggesting that the evolutionary adaptation of the lineage to utilize MK occurred prior to the divergence of these two related families. Members of the GoM-Arc-1 clade possess Mcr-like complexes (Fig. S3) and are suggested to use short-chain alkanes, possibly ethane (53, 54). Interestingly, the FpoMNO subunits of the ASW-3 MAG cluster with those of the other members of the Methanoperedenaceae family, while their FpoABCDHIJ1J2KL subunits are most similar to those of the ANME-2a and other members of the Methanosarcinales (Fig. S6). While the genes involved in MP biosynthesis are not known, the absence of the MK biosynthesis pathway indicate that ASW-3 likely uses MP. As the most basal lineage of this family, ASW-3 may have adapted to use MP after the evolutionary divergence of the GoM-Arc-I and Methanoperedenaceae, although further genomic representation of this lineage is required to verify this hypothesis.
Comparative genomic analyses of the Methanoperedenaceae MAGs revealed that none of these genomes encode an Rnf complex, which is hypothesized to reoxidize ferredoxin coupled to the transport of sodium ions out of the cell and the reduction of MP in marine ANME-2a (7, 55) and other methylotrophic methanogens (41, 56, 57). In the absence of this complex, ferredoxins may be reoxidized with a “truncated” Fpo complex, similar to the Fpo complex possessed by Methanosaeta thermophila (58). Alternatively, an electron-confurcating mechanism may be used for the reoxidation of ferredoxin, coenzyme M, and coenzyme B, coupled to the reduction of two F420 molecules via a cytoplasmic complex composed of a heterodisulfide reductase (HdrABC) and a F420 hydrogenase subunit B (FrhB) (24). The two additional F420H2 molecules may subsequently be fed back into the Fpo complex, greatly increasing the overall bioenergetic yield (24) (Fig. 2). All of the Methanoperedenaceae MAGs have the genetic potential for these alternate strategies for reoxidation of ferredoxin during AOM; however, further experimental validation is required to test these hypotheses.
Conservation of unique menaquinone: cytochrome c oxidoreductases within the Methanoperedenaceae.
Five different putative MK:cytochrome c oxidoreductase gene clusters (Fig. 1 and 2; Data Set S1A) that are hypothesized to mediate the transfer of electrons out of the cytoplasmic membrane were identified in the Methanoperedenaceae MAGs. These gene clusters include a noncanonical bc1/b6f complex adjacent to two hypothetical proteins and two multiheme (6 hemes) c-type cytochromes (MHCs; group 1), two clusters where a b-type cytochrome is adjacent to a 6-heme MHC (groups 2 and 3), and another two clusters where an NrfD-like transmembrane protein is adjacent to an electron-transferring 4Fe-4S ferredoxin iron-sulfur protein and MHCs (groups 4 and 5) (Fig. 2). These bc and NrfD complexes are frequently found in other metal-reducing microorganisms and mediate electron transport from the cytoplasm to the periplasm (59–61).
Most of the 16 Methanoperedenaceae MAGs (except CMD-1 and ASW-3) have more than one of these MK:cytochrome oxidoreductase complexes, and 10 have at least four (Fig. 1). ASW-3 is the only MAG not to encode any MK:cytochrome c oxidoreductases, which is consistent with its putative use of MP. A gene encoding a cytochrome b found to be most similar to that of “Ca. Methanohalarchaeum thermophilum” was identified in ASW-3; however, in the absence of a collocated MHC gene, the extracellular electron transfer step for this microorganism is unclear.
Phylogenetic analysis of the membrane-bound subunits of the MK:cytochrome c oxidoreductases (Fig. 2), which include the NrfD subunits (from groups 1 and 2) and the b-type cytochromes (from groups 3, 4, and 5), showed that they have been potentially laterally transferred from diverse donors (Fig. S7). The Methanoperedenaceae NrfD subunits formed independent clusters with sequences from members of the Dehalococcoidales family RBG-16-60-22 (group 1) and a single MAG (RBG-16-55-9) from the candidate phylum Bipolaricaulota (group 2) (Fig. S7A). The b-type cytochromes of the Methanoperedenaceae belong to three distinct clades (Fig. S7B). The b-type cytochromes from groups 3 and 4 clustered with proteins from GoM-ArcI, indicating vertical genetic inheritance from an ancestor of these two families, and group 5 proteins clustered with those from the class Archaeoglobi (40).
Phylogenetic analysis of the subunits of the MK:cytochrome oxidoreductases annotated in the Methanoperedenaceae MAGs. (A) Analysis of the NrfD subunits; (B) snalysis of the b-type cytochromes. Bootstrap values for the maximum-likelihood trees were determined using nonparametric bootstrapping with 100 replicates. The scale bars represent amino acid changes. Download FIG S7, PDF file, 0.7 MB (700.7KB, pdf) .
Copyright © 2020 Leu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The conservation of multiple conserved laterally transferred MK:cytochrome c oxidoreductases in most of the Methanoperedenaceae MAGs may contribute to the reported ability of members of the family to reduce a variety of electron acceptors with a range of redox potentials that include Fe(III) oxide reduction (–100mV to 100 mV) (62), nitrate (+433 mV) (24), and Mn(IV) (+380 mV) (36). Transcriptomic analyses have shown that different MK:cytochrome c oxidoreductases are expressed in different species of the genus “Ca. Methanoperedens” during AOM coupled to the reduction of Fe(III) oxides (9), Mn(IV) oxides (12), and nitrate (15, 24). A similar phenomenon has been observed for the species Geobacter sulfurreducens, where different extracellular electron pathways were used when different electron acceptors were reduced (63).
Potential electron acceptors used by the Methanoperedenaceae.
Annotation of the Methanoperedenaceae MAGs revealed a wide array of genes associated with previously undescribed respiratory strategies for the family that appear to have been acquired via LGT. Principally, these are putative terminal oxidoreductase complexes belonging to the complex-iron–sulfur–molybdenum (CISM) superfamily that were absent in the genomes of related archaeal lineages (Fig. 3). These complexes are composed of a catalytic subunit, an iron-sulfur protein, and a membrane-bound subunit and facilitate the transfer of electrons between the electron acceptor/donor and the MK pool (Fig. 2).
As previously reported, the MAGs M.Nitro, BLZ2, and IPS-1 encode respiratory nitrate reductases that are part of the CISM superfamily and have been demonstrated to mediate AOM coupled to nitrate reduction (15, 24, 29). Based on phylogenetic analysis (Fig. 3), genes encoding cytoplasmic nitrite oxidoreductases (NxrA) were identified in the IPS-1, BLZ2, and M.Nitro MAGs, and a nitrate reductase closely related to NarG proteins was identified in the BLZ2 MAG. Of the Methanoperedenaceae MAGs, only the M.Nitro and BLZ2 MAGs possess a putative nitrite reductase (NrfA) for DNRA. The M.Ferri MAG encodes an assimilatory nitrate reductase (NarB/NasA) most similar to a protein encoded by “Candidatus Magnetobacterium casensis” (Fig. 3). However, in the absence of an annotated nitrite reductase in the M.Ferri MAG, the potential of this microorganism for assimilatory nitrate reduction is unclear.
Multiple MAGs (ASW-2,3,5,6 and Mn-2) were also found to encode putative selenate reductases (SrdA) (Fig. 3), suggesting their ability for Se(VI)-dependent AOM. Recently, a bioreactor enrichment of a member of the genus “Ca. Methanoperedens” exhibited AOM activity when nitrate was replaced with selenate (26). However, as no meta-omic analyses was conducted for the community, it is unclear if the dominant “Ca. Methanoperedens” possessed a putative selenate reductase or if it was directly responsible for the observed selenate reduction.
The ASW-1 and ASW-3 MAGs encode a putative sulfur reductase (SreABC). This annotation is supported by its phylogenetic clustering of the catalytic subunit with SreA from Aquifex aeolicus (Fig. 3), which has been shown to reduce elemental sulfur, as well as tetrathionate and polysulfide (64). This is the first genomic evidence suggesting that members of the Methanoperedenaceae may be involved in respiratory sulfur-dependent AOM and warrants further investigation. ANME-1 has been proposed to couple AOM to the reduction of polysulfide in a biogenic hydrocarbon seep sediment, but this was based on the annotation and high expression of a putative sulfide: quinone oxidoreductase (SQR) (65). Genes for dissimilatory sulfate reduction pathways were absent in the Methanoperedenaceae MAGs, consistent with other ANME lineages (66). MGW-1 was recently speculated to directly couple AOM to sulfate reduction by utilizing assimilatory sulfate reduction pathways. This hypothesis was based on the lack of large MHCs or identifiable alternate electron acceptor complexes encoded in the MAG (30). Several of the Methanoperedenaceae MAGs, and those of other ANME lineages, contain candidate genes associated with assimilatory sulfate reduction, but a dissimilatory role for these has not been shown (66).
The M.Nitro MAG encodes two putative reductases belonging to the arsenate reductase (ArrA) and arsenite oxidase (ArxA) group (Fig. 3). The BLZ2, ASW-1, ASW-4, and IPS-1 MAGs also encode reductases that cluster with the M.Nitro ArxA-like sequence. The ArxA protein has been found to be capable of both arsenite oxidation and arsenate reduction (67), which would allow the Methanoperedenaceae possessing these ArxA-like proteins to utilize arsenate as a terminal electron acceptor. Proteins encoded by the ASW-3 and “Candidatus Acetothermum autotrophicum” (68) (Fig. 3) form a deep branching clade adjacent to the ArxA and ArrA groups, suggesting that these species might also have the potential to respire on arsenic compounds. It is noteworthy that the ASW-1, -3, and -4 MAGs were recovered from a Bangladesh arsenic-contaminated groundwater sample (Table 1), indicating a role for LGT in their niche-specific adaptation. The possibility of AOM coupled to arsenate [As(V)] reduction has important environmental implications given the wide distribution of arsenic in nature, including subsurface drinking water aquifers (69), and the toxicity and mobility of its reduced form, arsenite [As(III)] (70, 71). Arsenic reduction and mobilization have been linked to an inflow of organic carbon in contaminated aquifers where methane (∼1 mM) and arsenate cooccur (72, 73).
Additional putative oxidoreductase clades that are not closely associated with any well-characterized CISM proteins were also found in the Methanoperedenaceae MAGs. This includes two proteins encoded by the ASW-3 and ASW-6 MAGs that cluster with a protein of unknown function from a “Candidatus Brocadiales” MAG (74) and the CMD-1 protein that clusters with a protein from “Candidatus Brocadia fulgida,” an ammonium-oxidizing and nitrite-reducing microorganism (75). In general, given the large range of substrates utilized by the CISM superfamily and the few biochemically characterized proteins, the predicted functions of all those annotated in the Methanoperedenaceae require empirical verification. Nonetheless, the range of putative CISM superfamily proteins encoded by members of the family likely indicates diverse respiratory strategies that remain to be characterized.
Diversity of the MHCs in the Methanoperedenaceae.
Members of the Methanoperedenaceae possess a diverse repertoire of MHCs which have been suggested to facilitate the transfer of electrons from the reoxidation of MK to metal oxides (9, 10, 76) or direct interspecies electron transfer (DIET) to a syntrophic partner. Analyses of the Methanoperedenaceae revealed that they possess between 3 (MGW-1) and 49 (IPS-1) MHCs (containing at least three CXXCH motifs), with an average of 26, the highest average of any archaeal family (Data Sets S1F and S1G). Notably, relatively high numbers of MHCs per genome are almost exclusively found in microorganisms associated with DIET, metal and/or sulfur reduction, such as the Geobacteraceae (77) (≤87 MHCs), Shewanellaceae (78) (≤63 MHCs), Desulfurivibrionaceae (20), Desulfuromonadaceae (20), and Defferisomataceae (79) (≤50 MHCs) (Data Set S1G). Interestingly, 7 of the 16 members of the Methanoperedenaceae encode MHCs with more than 50 heme binding sites (ASW-5, ASW-6, BLZ2, HGW-1, M.ferri, Mn-1, and Mn-2), with the 113-heme MHC encoded by Mn-2 being the largest identified in any microorganism (Data Set S1F).
The 414 putative MHCs identified in the Methanoperedenaceae MAGs clustered into 82 orthologous protein families (Fig. S8). Only one protein family (OG0000252) included at least one MHC from each member, which suggests low conservation of these genes within the Methanoperedenaceae. Out of the 82 MHC protein families, 14 were identified in at least eight of the 16 MAGs, with five of these found within the conserved MK:cytochrome c oxidoreductase clusters. A lack of conservation of MHCs is also observed for anaerobic metal-respiring genus Geobacter organisms, where 14% of the MHCs encoded in six analyzed genomes were found to be conserved (60). Thirty-nine of the 82 MHC protein families had significant hits (1e−20, ≥50% AAI) to homologs from diverse lineages across the bacterial and archaeal domains in the GTDB89 database, indicating potential LGT of these genes (Fig. S9). These lineages notably included the metal-reducing Geobacteraceae and Shewanellaceae, along with the alkane-oxidizing Archaeoglobaceae, Methylomirabilota (NC10), and other ANME lineages (Fig. S9).
Abundance profiles for the MHC-orthologous protein families annotated in the Methanoperedenaceae MAGs. Download FIG S8, PDF file, 0.4 MB (458KB, pdf) .
Copyright © 2020 Leu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Network analysis of MHC-orthologous protein families in Methanoperedenaceae. Each cluster represents related MHCs. The color of the nodes represents the taxonomic lineage based on GTDB classification. The size of the nodes represents the number of CXXCH heme binding motifs identified in the proteins. The thicknesses of the lines represent amino acid identity between the two nodes. The shaded boxes represent the orthologous protein families. Download FIG S9, PDF file, 0.5 MB (483.2KB, pdf) .
Copyright © 2020 Leu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Putative functions of MHCs in the Methanoperedenaceae.
Very few of the Methanoperedenaceae MHCs could be associated with a specific function. Two orthologous groups were annotated as nitrite: ammonium oxidoreductases (NrfA), with homologs identified in bacterial MAGs classified as Anaerolineales (OG0004545; ≥66.3% AAI), and the candidate phylum UBP4 (OG0012490, 64.56% AAI). Several MHCs were also identified as part of the MK:cytochrome c oxidoreductase clusters, with homologs observed in members of the archaeal family Archaeoglobaceae (OG001557, OG000137, OG0001550; ≥57.3% AAI) (Fig. S9). MHC/S-layer fusion proteins were suggested to mediate the transfer of electrons across the S-layer for marine ANME-2 (7) and were relatively highly expressed by “Ca. Methanoperedens manganicus” and “Ca. Methanoperedens manganireducens” during AOM coupled to Mn(IV) reduction (12). Conversely, only low expression of MHC/S-layer protein genes borne by “Ca. Methanoperedens ferrireducens” was observed during AOM coupled to Fe(III) reduction (9). In addition, despite all the Methanoperedenaceae MAGs containing S-layer proteins, five do not encode MHC proteins with an S-layer domain (ASW-3, CMD-1, CMD-2, HGW-1, and MGW-1), indicating alternative mechanisms for electron transfer across the S-layer to extracellular MHCs for these species.
Predicted extracellular MHCs are hypothesized to facilitate the final transfer of electrons from the Methanoperedenaceae to metal oxides (9). Interestingly, “Ca. Methanoperedens manganicus” and “Ca. Methanoperedens manganireducens” showed differential expression patterns in the complement of shared extracellular MHCs during AOM coupled to Mn(IV) reduction. In addition, no orthologs for the two MHCs highly transcribed by “Ca. Methanoperedens ferrireducens” during AOM coupled to Fe(III) reduction (9) were identified in other members of the Methanoperedenaceae (OG0011636 and OG0003254) (Fig. S8), suggesting that BLZ2 utilizes a different MHC for iron reduction linked to AOM (10). These observations suggest that the Methanoperedenaceae can utilize multiple mechanisms for the reduction of similar metal oxides. Differential expression of conserved MHCs linked to extracellular electron transfer was also observed for different Geobacteraceae species enriched on electrodes when they were exposed to the same surface redox potential (80). As suggested for members of the Geobacteraceae, the large MHC repertoire possessed by the Methanoperedenaceae may enable adaptation to the use of a range of terminal electron acceptors.
This study has substantially improved the genome coverage of the Methanoperedenaceae. Comparative genomic analysis of this lineage highlights a metabolic plasticity not found in other ANME clades. The subsequent ability of members of the family to adapt to the use of terminal electron acceptors across a range of redox potentials likely contributes to their success in diverse environments (Table 1). Notably, based on the genome tree (Fig. 1) and the lack of conservation of MHCs (Fig. S8), the acquisition of these genes is not congruent with the genome-based phylogeny of the family, suggesting niche-specific adaptations as the main driver for these LGT events. While further studies are necessary to verify the general physiology and energy conservation mechanisms of the Methanoperedenaceae in different environments, this study provides genomic evidence that members of the family may play key roles in coupling cycling of carbon with selenate, sulfur, and arsenic in addition to nitrate and metal oxides. Continued sequencing and characterization of this lineage will reveal the full extent of their metabolic versatility and influence on global biogeochemical cycles.
MATERIALS AND METHODS
Recovery of the genomes from SRA.
The NCBI sequence read archive (SRA [81]) was accessed on the 22nd of March 2017, and 14,516 data sets classified as environmental metagenomes were downloaded. The metagenomic data sets were screened using GraftM (32) to search for 16S rRNA and mcrA gene sequences similar to those from members of the Methanoperedenaceae. For data sets for which members of the family were detected, all paired-end read sets were trimmed and quality filtered using PEAT v1.2.4 (82). For genomes, CMD-1 and CMD-2 and NCBI accession number SRR5161805 and SRR5161795 reads were coassembled using metaSPAdes version 3.10.0 using the default parameters (83). For the ASW genomes, NCBI accession number SRR1563167, SRR1564103, SRR1573565, and SRR1573578 reads were coassembled using metaSPAdes version 3.10.0, with default parameters (83). Mapping of quality reads was performed using BamM v1.7.3, with default parameters (https://github.com/Ecogenomics/BamM). Metagenomic assembled genomes were recovered from the assembled metagenomes using uniteM v0.0.14 (https://github.com/dparks1134/UniteM). The Methanoperedenaceae MAGs were further refined by reassembling the mapped quality trimmed reads with SPAdes using the –careful and –trusted contig settings. Additional scaffolding and resolving ambiguous bases of the MAGs was performed using the “roundup” mode of FinishM, v0.0.7 (https://github.com/wwood/finishm). The completeness and contamination rates of the population bins were assessed using CheckM v1.0.11 (33) with the “lineage wf” command.
Functional annotation.
For all MAGs, open reading frames (ORFs) were called and annotated using Prokka v.1.12 (84). Additional annotation was performed using the blastp “verysensitive” setting in Diamond v0.9.18 (https://github.com/bbuchfink/diamond.git) against UniRef100 (accessed September 2017) (85), clusters of orthologous groups (COG) (86), Pfam 31 (87), and TIGRfam (released January 2014) (88). ORFs were also diamond blastp searched against Uniref100 (accessed September 2017) containing proteins with KO identifiers (IDs). The top hit for each gene with an E value of <1e−3 was mapped to the KO database (89) using the UniProt ID mapping files. Genes of interest were further verified using the NCBI’s conserved domain search to identify a conserved motif(s) present within the gene (90). Psortb v3.0 (91) was used to predict subcellular localization of the putative proteins. Pred-Tat was used to predict putative signal peptides (92). Putative MHCs were identified by ORFs possessing ≥3 CXXCH motifs. Putative MHCs were subsequently searched for cytochrome c-type protein domains using hmmsearch (HMMER v.3.1) (93) with PfamA (94).
Construction of genome trees.
The archaeal genome tree was constructed using GTDB-Tk (GTDBtk v0.2.2; https://github.com/Ecogenomics/GTDBTk/releases) with a concatenated set of 122 archaeon-specific conserved marker genes inferred from genomes available in the NCBI database (NCBI RefSeq release 83) (34). Marker genes were identified and aligned in each genome using HMMER v.3.1 (93) and concatenated, and trees were constructed using FastTree v.2.1.8 (95) with the WAG+GAMMA models. Support values were determined using 100 nonparametric bootstrapping with GenomeTreeTK. The trees were visualized using ARB (96) and formatted using Adobe Illustrator (Adobe, USA).
Construction of 16S rRNA gene tree.
The 16S rRNA gene was identified in MAGs and used to infer taxonomic assignment of the population genome implementing the SILVA 16S rRNA gene database (version 132). Sequences were aligned with 426 16S rRNA gene sequences retrieved from the SILVA database using SSU-align v0.1 (97). The phylogenetic tree was constructed using FastTree v2.1.8 (95) with the Generalised Time-Reversible and GAMMA models. Support values were determined using 100 nonparametric bootstrapping. The trees were visualized using ARB (96) and formatted using Adobe Illustrator.
Calculation of amino acid identity.
The Methanoperedenaceae MAGs identified in this study were compared to publicly available genomes of the family. Average amino acid identity (AAI) between the genomes was calculated using orthologous genes identified through reciprocal best BLAST hits using compareM v0.0.5 (https://github.com/dparks1134/CompareM).
Identification of orthologous proteins.
Homologous proteins across MAGs of all the Methanoperedenaceae, GoM-Arc I, ANME-2a, and ANME-2c were identified with OrthoFinder (98) v2.3.3 using default parameters. Gene counts of orthologous groups containing MHCs were used as input for a heatmap using the pheatmap package in R, and hierarchical clustering was performed using ward.D2 (99).
Construction of gene trees.
Genes of interest in the Methanoperedenaceae MAGs were compared against proteins from the GTDB v83 database (34) using the genetreetk “blast” command to identify closely related sequences. For the generation of the gene tree for catalytic subunits of the CISM superfamily, curated protein sequences were also added in the analysis. Accession numbers and amino acid sequences are included in Data Set S1B. For the generation of the gene tree for the catalytic subunits of the group 1 and group 3 NiFe dehydrogenase, curated sequences from the work of Greening et al. (100) were included in the analysis. Accession numbers and amino acid sequences can be found in Data Set S1C. The sequences were subsequently aligned using mafft v7.221 (101) with the -auto function, and the alignment was trimmed using the trimal v1.2 (https://github.com/scapella/trimal) “-automated1” option. A phylogenetic tree was constructed using RAxML v8.2.9 (102) with the following parameters: raxmlHPC-PTHREADS-SSE3 -T 30 -m PROTGAMMALG -p 12345. Bootstrap values were calculated via nonparametric bootstrapping with 100 replicates. The trees were visualized using ARB (96) or iToL (103) and formatted using Adobe Illustrator (Adobe, USA).
Network analysis of MHCs.
Putative MHCs from the GTDB v89 database were identified by ORFs possessing ≥3 CXXCH motifs. Putative MHCs were subsequently searched for cytochrome c-type protein domains using hmmsearch (HMMER v.3.1) (93) with PfamA (94). Proteins from each Methanoperedenaceae orthogroup were subjected to a blast search against the GTDB v89 MHC protein database using DIAMOND with an E value cutoff of 1e–20 and ≥50% AAI. The result was visualized in Cytoscape v3.7.1, with clusters that contained only, or no, Methanoperedenaceae homologs removed.
Data availability.
The genomes assembled in this study have been deposited in the NCBI database under the accession numbers SAMN10961276 to SAMN10961283.
ACKNOWLEDGMENTS
This work was supported by the Australian Research Council (ARC) (grant FT170100070) and the U.S. Department of Energy’s Office of Biological Environmental Research (grant DE-SC0016469). A.O.L. was supported by an ARC Australian Postgraduate Award, and S.J.M. was partly supported by an ARC Future Fellowship (FT190100211).
We thank the AWMC team, particularly Shihu Hu and Zhiguo Yuan, for their ongoing collaboration working on various “Ca. Methanoperedens” enrichments.
We have nothing to disclose.
Footnotes
Citation Leu AO, McIlroy SJ, Ye J, Parks DH, Orphan VJ, Tyson GW. 2020. Lateral gene transfer drives metabolic flexibility in the anaerobic methane-oxidizing archaeal family Methanoperedenaceae. mBio 11:e01325-20. https://doi.org/10.1128/mBio.01325-20.
REFERENCES
- 1.Reeburgh WS. 2007. Oceanic methane biogeochemistry. Chem Rev 107:486–513. doi: 10.1021/cr050362v. [DOI] [PubMed] [Google Scholar]
- 2.Segarra KEA, Schubotz F, Samarkin V, Yoshinaga MY, Hinrichs K-U, Joye SB. 2015. High rates of anaerobic methane oxidation in freshwater wetlands reduce potential atmospheric methane emissions. Nat Commun 6:7477. doi: 10.1038/ncomms8477. [DOI] [PubMed] [Google Scholar]
- 3.Martinez-Cruz K, Sepulveda-Jauregui A, Casper P, Anthony KW, Smemo KA, Thalasso F. 2018. Ubiquitous and significant anaerobic oxidation of methane in freshwater lake sediments. Water Res 144:332–340. doi: 10.1016/j.watres.2018.07.053. [DOI] [PubMed] [Google Scholar]
- 4.Thamdrup B, Steinsdóttir HGR, Bertagnolli AD, Padilla CC, Patin NV, Garcia‐Robledo E, Bristow LA, Stewart FJ. 2019. Anaerobic methane oxidation is an important sink for methane in the ocean’s largest oxygen minimum zone. Limnol Oceanogr 64:2569–2585. doi: 10.1002/lno.11235. [DOI] [Google Scholar]
- 5.Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MMM, Schreiber F, Dutilh BE, Zedelius J, de Beer D, Gloerich J, Wessels HJCT, van Alen T, Luesken F, Wu ML, van de Pas-Schoonen KT, Op den Camp HJM, Janssen-Megens EM, Francoijs K-J, Stunnenberg H, Weissenbach J, Jetten MSM, Strous M. 2010. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464:543–548. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- 6.McGlynn SE. 2017. Energy metabolism during anaerobic methane oxidation in ANME archaea. Microbes Environ 32:5–13. doi: 10.1264/jsme2.ME16166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGlynn SE, Chadwick GL, Kempes CP, Orphan VJ. 2015. Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature 526:531–535. doi: 10.1038/nature15512. [DOI] [PubMed] [Google Scholar]
- 8.Wegener G, Krukenberg V, Riedel D, Tegetmeyer HE, Boetius A. 2015. Intercellular wiring enables electron transfer between methanotrophic archaea and bacteria. Nature 526:587–590. doi: 10.1038/nature15733. [DOI] [PubMed] [Google Scholar]
- 9.Cai C, Leu AO, Xie G-J, Guo J, Feng Y, Zhao J-X, Tyson GW, Yuan Z, Hu S. 2018. A methanotrophic archaeon couples anaerobic oxidation of methane to Fe (III) reduction. ISME J 12:1929–1939. doi: 10.1038/s41396-018-0109-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ettwig KF, Zhu B, Speth D, Keltjens JT, Jetten MSM, Kartal B. 2016. Archaea catalyze iron-dependent anaerobic oxidation of methane. Proc Natl Acad Sci U S A 113:12792–12796. doi: 10.1073/pnas.1609534113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scheller S, Yu H, Chadwick GL, McGlynn SE, Orphan VJ. 2016. Artificial electron acceptors decouple archaeal methane oxidation from sulfate reduction. Science 351:703–707. doi: 10.1126/science.aad7154. [DOI] [PubMed] [Google Scholar]
- 12.Leu AO, Cai C, McIlroy SJ, Southam G, Orphan VJ, Yuan Z, Hu S, Tyson GW. 2020. Anaerobic methane oxidation coupled to manganese reduction by members of the Methanoperedenaceae. ISME J 14:1030–1041. doi: 10.1038/s41396-020-0590-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knittel K, Lösekann T, Boetius A, Kort R, Amann R. 2005. Diversity and distribution of methanotrophic archaea at cold seeps. Appl Environ Microbiol 71:467–479. doi: 10.1128/AEM.71.1.467-479.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orphan VJ, House CH, Hinrichs K-U, McKeegan KD, DeLong EF. 2002. Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc Natl Acad Sci U S A 99:7663–7668. doi: 10.1073/pnas.072210299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haroon MF, Hu S, Shi Y, Imelfort M, Keller J, Hugenholtz P, Yuan Z, Tyson GW. 2013. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500:567–570. doi: 10.1038/nature12375. [DOI] [PubMed] [Google Scholar]
- 16.Orphan VJ, Hinrichs KU, Ussler W, Paull CK, Taylor LT, Sylva SP, Hayes JM, Delong EF. 2001. Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl Environ Microbiol 67:1922–1934. doi: 10.1128/AEM.67.4.1922-1934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pernthaler A, Dekas AE, Brown CT, Goffredi SK, Embaye T, Orphan VJ. 2008. Diverse syntrophic partnerships from deep-sea methane vents revealed by direct cell capture and metagenomics. Proc Natl Acad Sci U S A 105:7052–7057. doi: 10.1073/pnas.0711303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatzenpichler R, Connon SA, Goudeau D, Malmstrom RR, Woyke T, Orphan VJ. 2016. Visualizing in situ translational activity for identifying and sorting slow-growing archaeal-bacterial consortia. Proc Natl Acad Sci U S A 113:E4069–E4078. doi: 10.1073/pnas.1603757113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schreiber L, Holler T, Knittel K, Meyerdierks A, Amann R. 2010. Identification of the dominant sulfate-reducing bacterial partner of anaerobic methanotrophs of the ANME-2 clade. Environ Microbiol 12:2327–2340. doi: 10.1111/j.1462-2920.2010.02275.x. [DOI] [PubMed] [Google Scholar]
- 20.Skennerton CT, Chourey K, Iyer R, Hettich RL, Tyson GW, Orphan VJ. 2017. Methane-fueled syntrophy through extracellular electron transfer: uncovering the genomic traits conserved within diverse bacterial partners of anaerobic methanotrophic archaea. mBio 8:e00530-17. doi: 10.1128/mBio.00530-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holler T, Widdel F, Knittel K, Amann R, Kellermann MY, Hinrichs K-U, Teske A, Boetius A, Wegener G. 2011. Thermophilic anaerobic oxidation of methane by marine microbial consortia. ISME J 5:1946–1956. doi: 10.1038/ismej.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niemann H, Lösekann T, de Beer D, Elvert M, Nadalig T, Knittel K, Amann R, Sauter EJ, Schlüter M, Klages M, Foucher JP, Boetius A. 2006. Novel microbial communities of the Haakon Mosby mud volcano and their role as a methane sink. Nature 443:854–858. doi: 10.1038/nature05227. [DOI] [PubMed] [Google Scholar]
- 23.Su GY, Zopfi J, Yao HY, Steinle L, Niemann H, Lehmann MF. 2019. Manganese/iron-supported sulfate-dependent anaerobic oxidation of methane by archaea in lake sediments. Limnol Oceanogr 65:863–875. doi: 10.1002/lno.11354. [DOI] [Google Scholar]
- 24.Arshad A, Speth DR, de Graaf RM, den Camp HJO, Jetten MS, Welte CU. 2015. A metagenomics-based metabolic model of nitrate-dependent anaerobic oxidation of methane by Methanoperedens-like archaea. Front Microbiol 6:1423. doi: 10.3389/fmicb.2015.01423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu Y-Z, Fu L, Ding J, Ding Z-W, Li N, Zeng RJ. 2016. Cr(VI) reduction coupled with anaerobic oxidation of methane in a laboratory reactor. Water Res 102:445–452. doi: 10.1016/j.watres.2016.06.065. [DOI] [PubMed] [Google Scholar]
- 26.Luo J-H, Chen H, Hu S, Cai C, Yuan Z, Guo J. 2018. Microbial selenate reduction driven by a denitrifying anaerobic methane oxidation biofilm. Environ Sci Technol 52:4006–4012. doi: 10.1021/acs.est.7b05046. [DOI] [PubMed] [Google Scholar]
- 27.Shen LD, Ouyang L, Zhu YZ, Trimmer M. 2019. Active pathways of anaerobic methane oxidation across contrasting riverbeds. ISME J 13:752–766. doi: 10.1038/s41396-018-0302-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berger S, Frank J, Martins PD, Jetten MS, Welte CU. 2017. High-quality draft genome sequence of “Candidatus Methanoperedens sp.” strain BLZ2, a nitrate-reducing anaerobic methane-oxidizing archaeon enriched in an anoxic bioreactor. Genome Announc 5:e01159-17. doi: 10.1128/genomeA.01159-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaksmaa A, Guerrero-Cruz S, van Alen TA, Cremers G, Ettwig KF, Lüke C, Jetten MSM. 2017. Enrichment of anaerobic nitrate-dependent methanotrophic ‘Candidatus Methanoperedens nitroreducens’ archaea from an Italian paddy field soil. Appl Microbiol Biotechnol 101:7075–7084. doi: 10.1007/s00253-017-8416-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ino K, Hernsdorf AW, Konno U, Kouduka M, Yanagawa K, Kato S, Sunamura M, Hirota A, Togo YS, Ito K, Fukuda A, Iwatsuki T, Mizuno T, Komatsu DD, Tsunogai U, Ishimura T, Amano Y, Thomas BC, Banfield JF, Suzuki Y. 2018. Ecological and genomic profiling of anaerobic methane-oxidizing archaea in a deep granitic environment. ISME J 12:31–47. doi: 10.1038/ismej.2017.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hernsdorf AW, Amano Y, Miyakawa K, Ise K, Suzuki Y, Anantharaman K, Probst A, Burstein D, Thomas BC, Banfield JF. 2017. Potential for microbial H2 and metal transformations associated with novel bacteria and archaea in deep terrestrial subsurface sediments. ISME J 11:1915–1929. doi: 10.1038/ismej.2017.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boyd JA, Woodcroft BJ, Tyson GW. 2018. GraftM: a tool for scalable, phylogenetically informed classification of genes within metagenomes. Nucleic Acids Res 46:e59. doi: 10.1093/nar/gky174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil P-A, Hugenholtz P. 2018. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol 36:996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 35.Konstantinidis KT, Tiedje JM. 2005. Towards a genome-based taxonomy for prokaryotes. J Bacteriol 187:6258–6264. doi: 10.1128/JB.187.18.6258-6264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thauer RK, Jungermann K, Decker K. 1977. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev 41:100–180. doi: 10.1128/MMBR.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nauhaus K, Treude T, Boetius A, Kruger M. 2005. Environmental regulation of the anaerobic oxidation of methane: a comparison of ANME-I and ANME-II communities. Environ Microbiol 7:98–106. doi: 10.1111/j.1462-2920.2004.00669.x. [DOI] [PubMed] [Google Scholar]
- 38.Meyerdierks A, Kube M, Kostadinov I, Teeling H, Glöckner FO, Reinhardt R, Amann R. 2010. Metagenome and mRNA expression analyses of anaerobic methanotrophic archaea of the ANME-1 group. Environ Microbiol 12:422–439. doi: 10.1111/j.1462-2920.2009.02083.x. [DOI] [PubMed] [Google Scholar]
- 39.Loach PA. 1976. Oxidation-reduction potentials, absorbance bands and molar absorbance of compounds used in biochemical studies. Handb Biochem Mol Biol 1:122–130. [Google Scholar]
- 40.Jungbluth SP, Amend JP, Rappé MS. 2017. Metagenome sequencing and 98 microbial genomes from Juan de Fuca Ridge flank subsurface fluids. Sci Data 4:170037. doi: 10.1038/sdata.2017.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Welte C, Deppenmeier U. 2014. Bioenergetics and anaerobic respiratory chains of aceticlastic methanogens. Biochim Biophys Acta 1837:1130–1147. doi: 10.1016/j.bbabio.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 42.Kanai T, Matsuoka R, Beppu H, Nakajima A, Okada Y, Atomi H, Imanaka T. 2011. Distinct physiological roles of the three [NiFe]-hydrogenase orthologs in the hyperthermophilic archaeon Thermococcus kodakarensis. J Bacteriol 193:3109–3116. doi: 10.1128/JB.01072-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma K, Schicho RN, Kelly RM, Adams M. 1993. Hydrogenase of the hyperthermophile Pyrococcus furiosus is an elemental sulfur reductase or sulfhydrogenase: evidence for a sulfur-reducing hydrogenase ancestor. Proc Natl Acad Sci U S A 90:5341–5344. doi: 10.1073/pnas.90.11.5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berney M, Greening C, Hards K, Collins D, Cook GM. 2014. Three different [NiFe] hydrogenases confer metabolic flexibility in the obligate aerobe Mycobacterium smegmatis. Environ Microbiol 16:318–330. doi: 10.1111/1462-2920.12320. [DOI] [PubMed] [Google Scholar]
- 45.Stanley EH, Casson NJ, Christel ST, Crawford JT, Loken LC, Oliver SK. 2016. The ecology of methane in streams and rivers: patterns, controls, and global significance. Ecol Monogr 86:146–171. doi: 10.1890/15-1027. [DOI] [Google Scholar]
- 46.Deppenmeier U, Blaut M, Mahlmann A, Gottschalk G. 1990. Reduced coenzyme F420: heterodisulfide oxidoreductase, a proton-translocating redox system in methanogenic bacteria. Proc Natl Acad Sci U S A 87:9449–9453. doi: 10.1073/pnas.87.23.9449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Timmers PH, Welte CU, Koehorst JJ, Plugge CM, Jetten MS, Stams AJ. 2017. Reverse methanogenesis and respiration in methanotrophic archaea. Archaea 2017:1654237. doi: 10.1155/2017/1654237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tran QH, Unden G. 1998. Changes in the proton potential and the cellular energetics of Escherichia coli during growth by aerobic and anaerobic respiration or by fermentation. Eur J Biochem 251:538–543. doi: 10.1046/j.1432-1327.1998.2510538.x. [DOI] [PubMed] [Google Scholar]
- 49.Tietze M, Beuchle A, Lamla I, Orth N, Dehler M, Greiner G, Beifuss U. 2003. Redox potentials of methanophenazine and CoB-S-S-CoM, factors involved in electron transport in methanogenic archaea. Chembiochem 4:333–335. doi: 10.1002/cbic.200390053. [DOI] [PubMed] [Google Scholar]
- 50.Gonzalez O, Gronau S, Pfeiffer F, Mendoza E, Zimmer R, Oesterhelt D. 2009. Systems analysis of bioenergetics and growth of the extreme halophile Halobacterium salinarum. PLoS Comput Biol 5:e1000332. doi: 10.1371/journal.pcbi.1000332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jones AJ, Blaza JN, Varghese F, Hirst J. 2017. Respiratory complex I in Bos taurus and Paracoccus denitrificans pumps four protons across the membrane for every NADH oxidized. J Biol Chem 292:4987–4995. doi: 10.1074/jbc.M116.771899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sazanov LA. 2015. A giant molecular proton pump: structure and mechanism of respiratory complex I. Nat Rev Mol Cell Biol 16:375–388. doi: 10.1038/nrm3997. [DOI] [PubMed] [Google Scholar]
- 53.Dombrowski N, Seitz KW, Teske AP, Baker BJ. 2017. Genomic insights into potential interdependencies in microbial hydrocarbon and nutrient cycling in hydrothermal sediments. Microbiome 5:106. doi: 10.1186/s40168-017-0322-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Borrel G, Adam PS, McKay LJ, Chen L-X, Sierra-García IN, Sieber CMK, Letourneur Q, Ghozlane A, Andersen GL, Li W-J, Hallam SJ, Muyzer G, de Oliveira VM, Inskeep WP, Banfield JF, Gribaldo S. 2019. Wide diversity of methane and short-chain alkane metabolisms in uncultured archaea. Nat Microbiol 4:603–613. doi: 10.1038/s41564-019-0363-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang F-P, Zhang Y, Chen Y, He Y, Qi J, Hinrichs K-U, Zhang X-X, Xiao X, Boon N. 2014. Methanotrophic archaea possessing diverging methane-oxidizing and electron-transporting pathways. ISME J 8:1069–1078. doi: 10.1038/ismej.2013.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schlegel K, Muller V. 2013. Evolution of Na and H bioenergetics in methanogenic archaea. Biochem Soc Trans 41:421–426. doi: 10.1042/BST20120294. [DOI] [PubMed] [Google Scholar]
- 57.Schlegel K, Welte C, Deppenmeier U, Müller V. 2012. Electron transport during aceticlastic methanogenesis by Methanosarcina acetivorans involves a sodium-translocating Rnf complex. FEBS J 279:4444–4452. doi: 10.1111/febs.12031. [DOI] [PubMed] [Google Scholar]
- 58.Welte C, Deppenmeier U. 2011. Membrane-bound electron transport in Methanosaeta thermophila. J Bacteriol 193:2868–2870. doi: 10.1128/JB.00162-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anderson I, Risso C, Holmes D, Lucas S, Copeland A, Lapidus A, Cheng J-F, Bruce D, Goodwin L, Pitluck S, Saunders E, Brettin T, Detter JC, Han C, Tapia R, Larimer F, Land M, Hauser L, Woyke T, Lovley D, Kyrpides N, Ivanova N. 2011. Complete genome sequence of Ferroglobus placidus AEDII12DO. Stand Genomic Sci 5:50–60. doi: 10.4056/sigs.2225018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Butler JE, Young ND, Lovley DR. 2010. Evolution of electron transfer out of the cell: comparative genomics of six Geobacter genomes. BMC Genomics 11:40. doi: 10.1186/1471-2164-11-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mardanov AV, Slododkina GB, Slobodkin AI, Beletsky AV, Gavrilov SN, Kublanov IV, Bonch-Osmolovskaya EA, Skryabin KG, Ravin NV. 2015. The Geoglobus acetivorans genome: Fe (III) reduction, acetate utilization, autotrophic growth, and degradation of aromatic compounds in a hyperthermophilic archaeon. Appl Environ Microbiol 81:1003–1012. doi: 10.1128/AEM.02705-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Straub KL, Schink B. 2004. Ferrihydrite reduction by Geobacter species is stimulated by secondary bacteria. Arch Microbiol 182:175–181. doi: 10.1007/s00203-004-0686-0. [DOI] [PubMed] [Google Scholar]
- 63.Levar CE, Hoffman CL, Dunshee AJ, Toner BM, Bond DR. 2017. Redox potential as a master variable controlling pathways of metal reduction by Geobacter sulfurreducens. ISME J 11:741–752. doi: 10.1038/ismej.2016.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guiral M, Tron P, Aubert C, Gloter A, Iobbi-Nivol C, Giudici-Orticoni M-T. 2005. A membrane-bound multienzyme, hydrogen-oxidizing, and sulfur-reducing complex from the hyperthermophilic bacterium Aquifex aeolicus. J Biol Chem 280:42004–42015. doi: 10.1074/jbc.M508034200. [DOI] [PubMed] [Google Scholar]
- 65.Vigneron A, Alsop EB, Cruaud P, Philibert G, King B, Baksmaty L, Lavallee D, Lomans BP, Eloe-Fadrosh E, Kyrpides NC, Head IM, Tsesmetzis N. 2019. Contrasting pathways for anaerobic methane oxidation in Gulf of Mexico cold seep sediments. mSystems 4:e00091-18. doi: 10.1128/mSystems.00091-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu H, Susanti D, McGlynn SE, Skennerton CT, Chourey K, Iyer R, Scheller S, Tavormina PL, Hettich RL, Mukhopadhyay B, Orphan VJ. 2018. Comparative genomics and proteomic analysis of assimilatory sulfate reduction pathways in anaerobic methanotrophic archaea. Front Microbiol 9:2917. doi: 10.3389/fmicb.2018.02917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zargar K, Conrad A, Bernick DL, Lowe TM, Stolc V, Hoeft S, Oremland RS, Stolz J, Saltikov CW. 2012. ArxA, a new clade of arsenite oxidase within the DMSO reductase family of molybdenum oxidoreductases. Environ Microbiol 14:1635–1645. doi: 10.1111/j.1462-2920.2012.02722.x. [DOI] [PubMed] [Google Scholar]
- 68.Takami H, Noguchi H, Takaki Y, Uchiyama I, Toyoda A, Nishi S, Chee G-J, Arai W, Nunoura T, Itoh T, Hattori M, Takai K. 2012. A deeply branching thermophilic bacterium with an ancient acetyl-CoA pathway dominates a subsurface ecosystem. PLoS One 7:e30559. doi: 10.1371/journal.pone.0030559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nordstrom DK. 2002. Worldwide occurrences of arsenic in ground water. Science 296:2143–2145. doi: 10.1126/science.1072375. [DOI] [PubMed] [Google Scholar]
- 70.Smedley PL, Kinniburgh D. 2002. A review of the source, behaviour and distribution of arsenic in natural waters. Appl Geochem 17:517–568. doi: 10.1016/S0883-2927(02)00018-5. [DOI] [Google Scholar]
- 71.Council NR. 1999. Arsenic in drinking water. National Academies Press, Washington, DC. [PubMed] [Google Scholar]
- 72.Harvey CF, Swartz CH, Badruzzaman ABM, Keon-Blute N, Yu W, Ali MA, Jay J, Beckie R, Niedan V, Brabander D, Oates PM, Ashfaque KN, Islam S, Hemond HF, Ahmed MF. 2002. Arsenic mobility and groundwater extraction in Bangladesh. Science 298:1602–1606. doi: 10.1126/science.1076978. [DOI] [PubMed] [Google Scholar]
- 73.Polizzotto ML, Harvey CF, Sutton SR, Fendorf S. 2005. Processes conducive to the release and transport of arsenic into aquifers of Bangladesh. Proc Natl Acad Sci U S A 102:18819–18823. doi: 10.1073/pnas.0509539103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Anantharaman K, Brown CT, Hug LA, Sharon I, Castelle CJ, Probst AJ, Thomas BC, Singh A, Wilkins MJ, Karaoz U, Brodie EL, Williams KH, Hubbard SS, Banfield JF. 2016. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat Commun 7:13219. doi: 10.1038/ncomms13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gori F, Tringe SG, Kartal B, Marchiori E, Machiori E, Jetten MSM. 2011. The metagenomic basis of anammox metabolism in Candidatus ‘Brocadia fulgida.’ Biochem Soc Trans 39:1799–1804. doi: 10.1042/BST20110707. [DOI] [PubMed] [Google Scholar]
- 76.Kletzin A, Heimerl T, Flechsler J, van Niftrik L, Rachel R, Klingl A. 2015. Cytochromes c in Archaea: distribution, maturation, cell architecture, and the special case of Ignicoccus hospitalis. Front Microbiol 6:439. doi: 10.3389/fmicb.2015.00439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Methé BA, Nelson KE, Eisen JA, Paulsen IT, Nelson W, Heidelberg JF, Wu D, Wu M, Ward N, Beanan MJ, Dodson RJ, Madupu R, Brinkac LM, Daugherty SC, DeBoy RT, Durkin AS, Gwinn M, Kolonay JF, Sullivan SA, Haft DH, Selengut J, Davidsen TM, Zafar N, White O, Tran B, Romero C, Forberger HA, Weidman J, Khouri H, Feldblyum TV, Utterback TR, Van Aken SE, Lovley DR, Fraser CM. 2003. Genome of Geobacter sulfurreducens: metal reduction in subsurface environments. Science 302:1967–1969. doi: 10.1126/science.1088727. [DOI] [PubMed] [Google Scholar]
- 78.Heidelberg JF, Paulsen IT, Nelson KE, Gaidos EJ, Nelson WC, Read TD, Eisen JA, Seshadri R, Ward N, Methe B, Clayton RA, Meyer T, Tsapin A, Scott J, Beanan M, Brinkac L, Daugherty S, DeBoy RT, Dodson RJ, Durkin AS, Haft DH, Kolonay JF, Madupu R, Peterson JD, Umayam LA, White O, Wolf AM, Vamathevan J, Weidman J, Impraim M, Lee K, Berry K, Lee C, Mueller J, Khouri H, Gill J, Utterback TR, McDonald LA, Feldblyum TV, Smith HO, Venter JC, Nealson KH, Fraser CM. 2002. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat Biotechnol 20:1118–1123. doi: 10.1038/nbt749. [DOI] [PubMed] [Google Scholar]
- 79.Slobodkina GB, Reysenbach A-L, Panteleeva AN, Kostrikina NA, Wagner ID, Bonch-Osmolovskaya EA, Slobodkin AI. 2012. Deferrisoma camini gen. nov., sp. nov., a moderately thermophilic, dissimilatory iron (III)-reducing bacterium from a deep-sea hydrothermal vent that forms a distinct phylogenetic branch in the Deltaproteobacteria. Int J Syst Evol Microbiol 62:2463–2468. doi: 10.1099/ijs.0.038372-0. [DOI] [PubMed] [Google Scholar]
- 80.Ishii S, Suzuki S, Tenney A, Nealson KH, Bretschger O. 2018. Comparative metatranscriptomics reveals extracellular electron transfer pathways conferring microbial adaptivity to surface redox potential changes. ISME J 12:2844–2863. doi: 10.1038/s41396-018-0238-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cochrane G, Karsch-Mizrachi I, Takagi T, International Nucleotide Sequence Database Collaboration. 2016. The International Nucleotide Sequence Database Collaboration. Nucleic Acids Res 44:D48–D50. doi: 10.1093/nar/gkv1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li Y-L, Weng J-C, Hsiao C-C, Chou M-T, Tseng C-W, Hung J-H editors. 2015. PEAT: an intelligent and efficient paired-end sequencing adapter trimming algorithm. BMC Bioinformatics 16:S2. doi: 10.1186/1471-2105-16-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res 27:824–116. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 85.Suzek BE, Huang H, McGarvey P, Mazumder R, Wu CH. 2007. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics 23:1282–1288. doi: 10.1093/bioinformatics/btm098. [DOI] [PubMed] [Google Scholar]
- 86.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D85. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Haft DH, Selengut JD, Richter RA, Harkins D, Basu MK, Beck E. 2013. TIGRFAMs and genome properties in 2013. Nucleic Acids Res 41:D387–D95. doi: 10.1093/nar/gks1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Marchler-Bauer A, Bryant SH. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res 32(Suppl 2):W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FSL. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bagos PG, Nikolaou EP, Liakopoulos TD, Tsirigos KD. 2010. Combined prediction of Tat and Sec signal peptides with hidden Markov models. Bioinformatics 26:2811–2817. doi: 10.1093/bioinformatics/btq530. [DOI] [PubMed] [Google Scholar]
- 93.Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol 7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, Studholme DJ, Yeats C, Eddy SR. 2004. The Pfam protein families database. Nucleic Acids Res 32(Suppl 1):D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Buchner A. 2004. ARB: a software environment for sequence data. Nucleic Acids Res 32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nawrocki EP, Kolbe DL, Eddy SR. 2009. Infernal 1.0: inference of RNA alignments. Bioinformatics 25:1335–1337. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Emms DM, Kelly S. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol 16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kolde R. 2019. Package ‘pheatmap.’ R Package. https://cran.r-project.org/web/packages/pheatmap/.
- 100.Greening C, Biswas A, Carere CR, Jackson CJ, Taylor MC, Stott MB, Cook GM, Morales SE. 2016. Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J 10:761–777. doi: 10.1038/ismej.2015.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Layton AC, Chauhan A, Williams DE, Mailloux B, Knappett PS, Ferguson AS, McKay LD, Alam MJ, Matin Ahmed K, van Geen A, Sayler GS. 2014. Metagenomes of microbial communities in arsenic- and pathogen-contaminated well and surface water from Bangladesh. Genome Announc 2:e01170-14. doi: 10.1128/genomeA.01170-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Medeiros JD, Leite LR, Pylro VS, Oliveira FS, Almeida VM, Fernandes GR, Salim ACM, Araujo FMG, Volpini AC, Oliveira G, Cuadros-Orellana S. 2017. Single-cell sequencing unveils the lifestyle and CRISPR-based population history of Hydrotalea sp. in acid mine drainage. Mol Ecol 26:5541–5551. doi: 10.1111/mec.14294. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Average amino acid identity (AAI%) for the Methanoperedenaceae genomes. AAI was calculated between each pair of genomes using CompareM. Download FIG S1, PDF file, 0.2 MB (172KB, pdf) .
Copyright © 2020 Leu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
16S rRNA gene-based phylogenetic placement of the Methanoperedenaceae MAGs. The 16S rRNA genes extracted from the Methanoperedenaceae MAGs from this study are indicated in red. Support values calculated via nonparametric bootstrapping. The scale bar represents numbers of changes per nucleotide position. Download FIG S2, PDF file, 0.5 MB (483.2KB, pdf) .
Copyright © 2020 Leu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phylogenetic analysis of methyl-coenzyme reductase subunit A (McrA). Putative genes recovered from the Methanoperedenaceae are indicated in red. The gene tree was inferred using maximum likelihood, and support values were calculated via nonparametric bootstrapping. The scale bar represents amino acid changes. Download FIG S3, PDF file, 0.1 MB (144.2KB, pdf) .
Copyright © 2020 Leu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sequences, identifiers, and statistics for genes used in the comparative analyses of the Methanoperedenaceae MAGs. (A) Genes encoding proteins involved in the methane oxidation pathway, energy conservation, and other metabolic pathways as shown in Fig. 2. (B) Amino acid sequences used in the CISM superfamily gene tree (Fig. 3). Amino acid sequences include curated sequences from Swiss-Prot and the work of Castelle et al. (C. J. Castelle, L. A. Hug, K. C. Wrighton, B. C. Thomas, et al., Nat Commun 4:2120, 2013, https://doi.org/10.1038/ncomms3120) and closely related sequences from GTDB r83 protein reference database (D. H. Parks, M. Chuvochina, D. W. Waite, C. Rinke, et al., Nat Biotechnol 36:996–1004, 2018, https://doi.org/10.1038/nbt.4229). (C) Amino acid sequences used in the catalytic subunits of the energy-converting NiFe hydrogenase. Amino acid sequences include curated sequences from Greening et al. (C. Greening, A. Biswas, C. R. Carere, C. J. Jackson, et al., ISME J 10:761–777, 2016, https://doi.org/10.1038/ismej.2015.153) and closely related sequences from the GTDB r83 protein reference database. (D) Genes encoding putative NiFe hydrogenase maturation proteins. (E) Best blastp hits of Fpo dehydrogenase subunits to the IMG database. Blastp hits show that divergent Fpo subunits are present in the Methanoperedenaceae MAGs, as seen in Fig. S6. Top blast hits to “Ca. Methanoperedens”-like protein sequences were excluded. (F) General statistic of multiheme c-type cytochromes (MHCs) in the ANME genomes. (G) MHC general statistics for all bacterial and archaeal families in the GTDB v89 database. Download Data Set S1, XLSX file, 0.2 MB (251.6KB, xlsx) .
Copyright © 2020 Leu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phylogenetic analysis of the subunits of the NiFe hydrogenases annotated in the Methanoperedenaceae genomes. (A) Analysis of the catalytic subunits of the energy-converting NiFe hydrogenases. (B) Analysis of the b-type cytochrome in the group 1 NiFe hydrogenases. Putative genes recovered from the Methanoperedenaceae are indicated in red. The gene trees were inferred using maximum likelihood and support values calculated via nonparametric bootstrapping. The reference sequences of group 1 and group 3 NiFe hydrogenases were acquired from the work of Greening et al. (C. Greening, A. Biswas, C. R. Carere, C. J. Jackson, et al., ISME J 10:761–777, 2016, https://doi.org/10.1038/ismej.2015.153) and the GTDB v83 reference sequences (D. H. Parks, M. Chuvochina, D. W. Waite, C. Rinke, et al., Nat Biotechnol 36:996–1004, 2018, https://doi.org/10.1038/nbt.4229). The scale bars represent amino acid changes. Download FIG S4, PDF file, 0.7 MB (688KB, pdf) .
Copyright © 2020 Leu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Subunit compositions of the Fpo dehydrogenase protein complexes and theoretical bioenergetics of energy metabolism in ANME-2a and Methanoperedenaceae. (A) Fpo subunit components for the ANME-2a and ASW-3 genomes (top left) and the other members of the Methanoperedenaceae (bottom left). The utilization of different electron carriers shows greater biochemical energetic gains based on more potential proton translocation. The colors orange and green depict Methanosarcinales-like and non-Methanosarcinales-like subunits. (B) Theoretical redox potential drop when utilizing MP (left) or MK (right) during F420H2 and Fd2– oxidation. This is due to differences between the membrane-bound electron carriers’ redox midpoint potential (Em) of –80 mV and –165 mV for MK and MP, respectively (M. Tietze, A. Beuchle, I. Lamla, N. Orth, et al., Chembiochem 4:333–335, 2003, https://doi.org/10.1002/cbic.200390053; Q. H. Tran and G. Unden, Eur J Biochem 251:538–543, 1998, https://doi.org/10.1046/j.1432-1327.1998.2510538.x). Download FIG S5, PDF file, 0.3 MB (348.2KB, pdf) .
Copyright © 2020 Leu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phylogenetic analysis of the Fpo subunits annotated in the Methanoperedenaceae genomes. (A) FpoA; (B) FpoB; (C) FpoC; (D) FpoD; (E) FpoH; (F) FpoI; (G) FpoJ1; (H) FpoJ2; (I) FpoK; (J) FpoL; (K) FpoM; (L) FpoN; (M) FpoO. Putative genes recovered from the Methanoperedenaceae are indicated in red. The gene trees were inferred using maximum likelihood and support values calculated via nonparametric bootstrapping. Reference genes and the taxonomy are from the GTDB v83 database (D. H. Parks, M. Chuvochina, D. W. Waite, C. Rinke, et al., Nat Biotechnol 36:996–1004, 2018, https://doi.org/10.1038/nbt.4229). Download FIG S6, PDF file, 2.1 MB (2.1MB, pdf) .
Copyright © 2020 Leu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phylogenetic analysis of the subunits of the MK:cytochrome oxidoreductases annotated in the Methanoperedenaceae MAGs. (A) Analysis of the NrfD subunits; (B) snalysis of the b-type cytochromes. Bootstrap values for the maximum-likelihood trees were determined using nonparametric bootstrapping with 100 replicates. The scale bars represent amino acid changes. Download FIG S7, PDF file, 0.7 MB (700.7KB, pdf) .
Copyright © 2020 Leu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Abundance profiles for the MHC-orthologous protein families annotated in the Methanoperedenaceae MAGs. Download FIG S8, PDF file, 0.4 MB (458KB, pdf) .
Copyright © 2020 Leu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Network analysis of MHC-orthologous protein families in Methanoperedenaceae. Each cluster represents related MHCs. The color of the nodes represents the taxonomic lineage based on GTDB classification. The size of the nodes represents the number of CXXCH heme binding motifs identified in the proteins. The thicknesses of the lines represent amino acid identity between the two nodes. The shaded boxes represent the orthologous protein families. Download FIG S9, PDF file, 0.5 MB (483.2KB, pdf) .
Copyright © 2020 Leu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The genomes assembled in this study have been deposited in the NCBI database under the accession numbers SAMN10961276 to SAMN10961283.