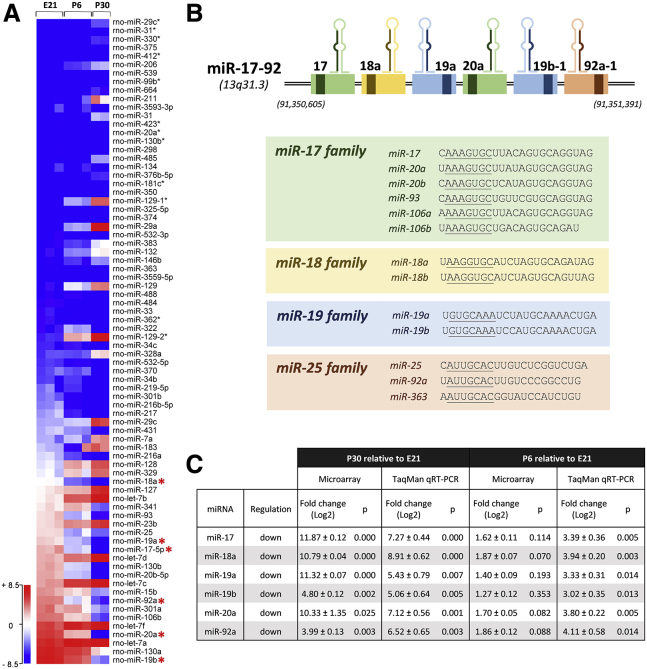
Figure 1.
miRNAs Are Differentially Expressed in RGCs during Development
(A) A representative heatmap of miRNA expression profiles constructed from a microarray analysis of retinal ganglion cells (RGCs) purified from embryonic day 21 (E21, n = 3 biological replicates), postnatal day 6 (P6, n = 3 biological replicates), and P30 (n = 2 biological replicates) Sprague-Dawley (SD) rats, showing 76 endogenously expressed miRNAs with significant differential expression (≥4-fold changes in expression levels) during development. Developmental ages (as biological replicates) are indicated in columns, and differentially expressed miRNAs are indicated in rows. All six members of the miR-17-92 cluster (red asterisks) were found to have significant downregulation from E21 to P30 (right panel). Blue (−8.5) and red (+8.5) in the color-coding scale represent relative low and high normalized miRNA expressions, respectively. A two-tailed moderated t test with a Benjamini-Hochberg correction for false discovery rate was used to compare the means of miRNA microarray expression values between E21 and P6 and between E21 and P30. A list of the 76 miRNAs can be found in Table S1, and all other microarray data that support the findings of this study have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO: GSE102458). (B) Illustration of the polycistronic miR-17-92 cluster region on human chromosome 13 (chromosome 15 for rats). Precursor transcripts derived from the miR-17-92 gene cluster contains six tandem stem-loop hairpin structures that yield six mature miRNAs: miR-17, miR-18a, miR-19a, miR-20a, miR-19b, and miR-92a. The miRNAs can be categorized into four miRNA families according to their conserved seed sequences (underlined). (C) Downregulation of the six miRNAs from E21 to P30 and from E21 to P6 was confirmed by TaqMan qRT-PCR (E21 vs. P6, miR-17-5p, p = 0.005; miR-18a, p = 0.003; miR-19a, p = 0.014; miR-19b, p = 0.013; miR-20a, p = 0.005; miR-92a, p = 0.014; E21 vs. P30, miR-17-5p, p = 0.0004; miR-18a, p = 0.001; miR-19a, p = 0.007; miR-19b, p = 0.005; miR-20a, p = 0.001; miR-92a, p = 0.003; n = 3 biological replicates for each age group). An unpaired two-tailed Student’s t test was used for TaqMan comparisons. All values are shown as mean ± SEM.