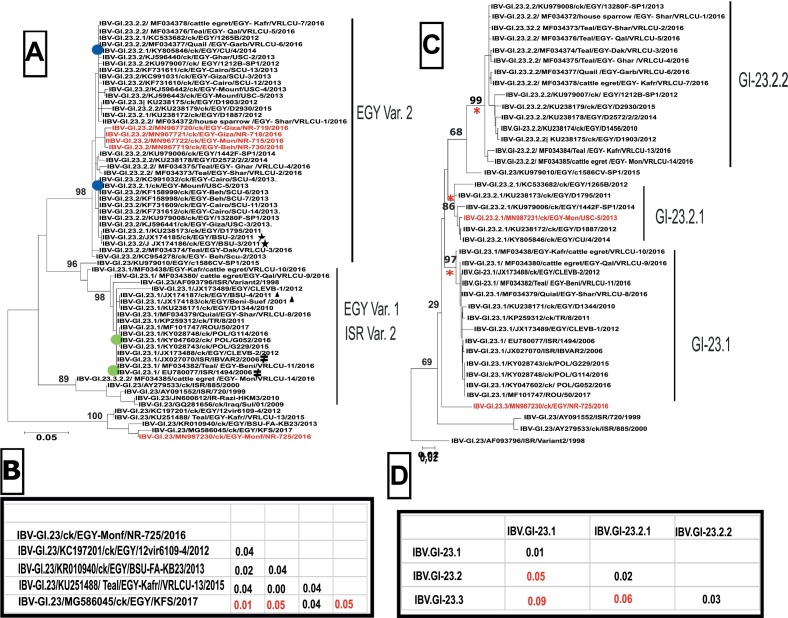
Fig. 1.
Phylogenetic analysis of IBV viruses within genotype GI-23 based on partial (a) and full (b) S1-gene sequences. The phyologenetic tree was generated using Maximum likelihood analysis for the partial S1 gene (290 nucleotides) including partial HVR3. Marked are reference strain for IS variant 2 viruses ( ), Egyptian variant 1 (
), Egyptian variant 1 ( ) and Egyptian variant 2 (
) and Egyptian variant 2 ( ). Prototype viruses used throughout the study include 2 EGY-var2 - (
). Prototype viruses used throughout the study include 2 EGY-var2 - ( ) and 2 EGY-var1-strains (
) and 2 EGY-var1-strains ( ). Virus from the small broiler flock (NR 725/16) and four additional viruses from the year 2016 are marked in red(A). In addition, the nucleotide distances between NR725/16 and related sequences in the gene bank are given (B). Phylogenetic analysis based on full S1 gene (1610 Nucleotide) resulted in establishing three sub-clades (GI-23.1, −23.2.1 and −23.2.2) (C). Viruses with full genome sequences in the study are highlighted in red. Distances between the proposed sub-clades are given below (D). Names include the genotype according to S1 gene, followed by accession number, species the virus was detected from with abbreviation for chicken (ck), country code according ISO 3166-1 alpha-3, laboratory identification number and year of publication.
). Virus from the small broiler flock (NR 725/16) and four additional viruses from the year 2016 are marked in red(A). In addition, the nucleotide distances between NR725/16 and related sequences in the gene bank are given (B). Phylogenetic analysis based on full S1 gene (1610 Nucleotide) resulted in establishing three sub-clades (GI-23.1, −23.2.1 and −23.2.2) (C). Viruses with full genome sequences in the study are highlighted in red. Distances between the proposed sub-clades are given below (D). Names include the genotype according to S1 gene, followed by accession number, species the virus was detected from with abbreviation for chicken (ck), country code according ISO 3166-1 alpha-3, laboratory identification number and year of publication.