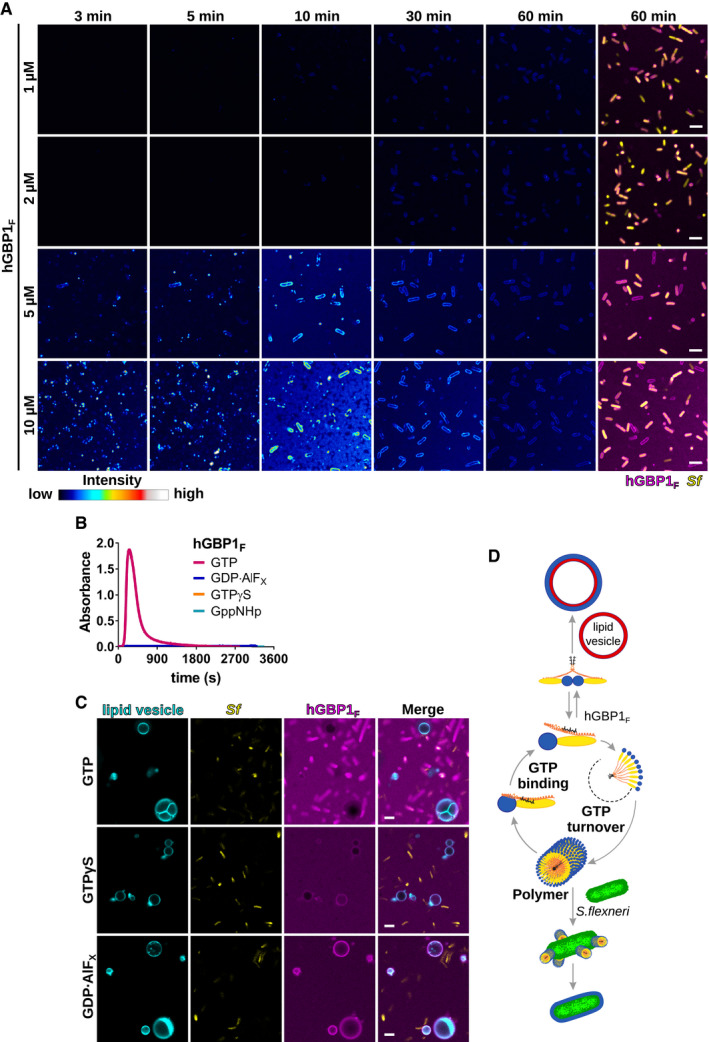
Figure EV1. Polymerizing hGBP1F binds directly to Shigella flexneri across a physiological range of hGBP1F protein concentrations.
-
ATime‐lapse microscopy frames of formaldehyde‐fixed GFP+ S. flexneri following admixture of varying concentrations of Alexa‐Fluor647‐labeled hGBP1F and 2 mM GTP. Individual time frames depict hGBP1F fluorescence intensity. Merged images of hGBP1F and S. flexneri fluorescence are shown for the 60 min time points.
-
BPolymerization of 10 μM hGBP1F monitored over time by absorption spectroscopy at 350 nm after addition of 2 mM (GTP, GppNHp, GTPγS) or 250 μM (GDP·AlFX) nucleotide.
-
CConfocal images taken of formaldehyde‐fixed GFP+ S. flexneri and rhodamine‐labeled lipid vesicles 20 min after addition of 10 μM Alexa‐Fluor647‐hGBP1F and 2 mM GTP, 2 mM GTPγS, or 250 μM GDP·AlFX.
-
DModel: In its GTP‐bound conformation, the C‐terminal farnesyl tail is released from its hydrophobic pocket allowing hGBP1 to bind to lipid vesicles in vitro and to host membranes inside human cells. Direct binding to bacteria on the other hand requires hGBP1 to self‐assemble over several GTP turnover cycles into polymers. These hGBP1 polymers bind to S. flexneri directly and then transition into a bacteria‐encapsulating protein coat.
