Figure 2. hGBP1 polymerization is required for bacterial binding.
-
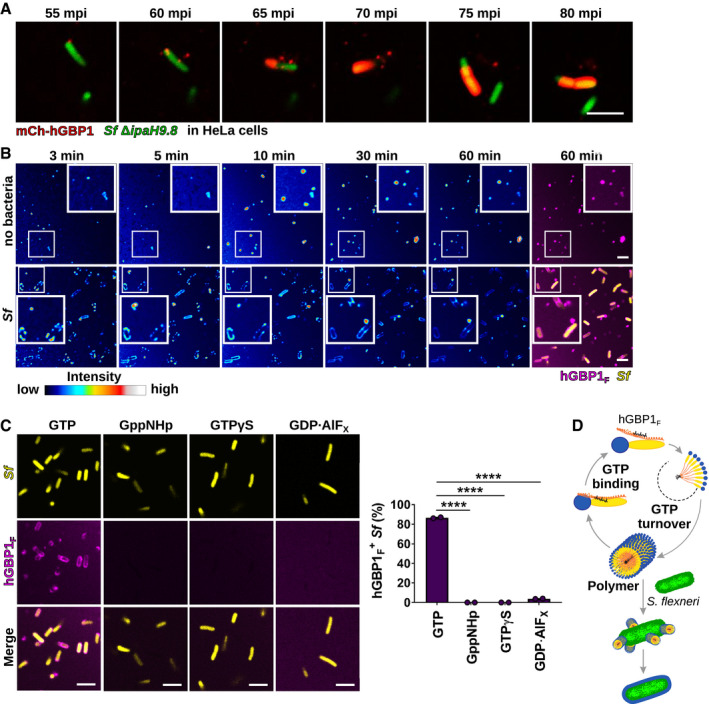
ATranslocation of ectopically expressed mCherry‐hGBP1 to cytosolic GFP+ Shigella flexneri ΔipaH9.8 in HeLa hGBP1‐KO cells was monitored by time‐lapse microscopy. Individual time frames of Movie EV1 starting at 55 min post‐infection (mpi) are shown.
-
BConfocal time‐lapse microscopy was used to image 10 μM Alexa‐Fluor647‐hGBP1F supplemented with 2 mM GTP in the presence or absence of formaldehyde‐fixed GFP+ S. flexneri. Individual time frames of Movie EV2 depict hGBP1F fluorescence intensity. Merged images of hGBP1F and S. flexneri fluorescence are shown for the 60 min time points.
-
CImages were taken at 45 min after addition of 10 μM Alexa‐Fluor647‐hGBP1F to formaldehyde‐fixed GFP+ S. flexneri in the presence of indicated nucleotides (GTP, natural substrate; GppNHp, non‐hydrolysable GTP analog; GTPγS, slowly hydrolysable GTP analog; GDP·AlFX, GTP transition state analog). hGBP1‐associated bacteria after 45 min were quantified. Combined data from two independent experiments are shown as mean ± SEM. Significance was determined by one‐way ANOVA with Tukey's multiple comparison test. ****P ≤ 0.0001.
-
DModel: hGBP1 polymers bind to S. flexneri directly and transition into a bacterium‐encapsulating protein coat.
