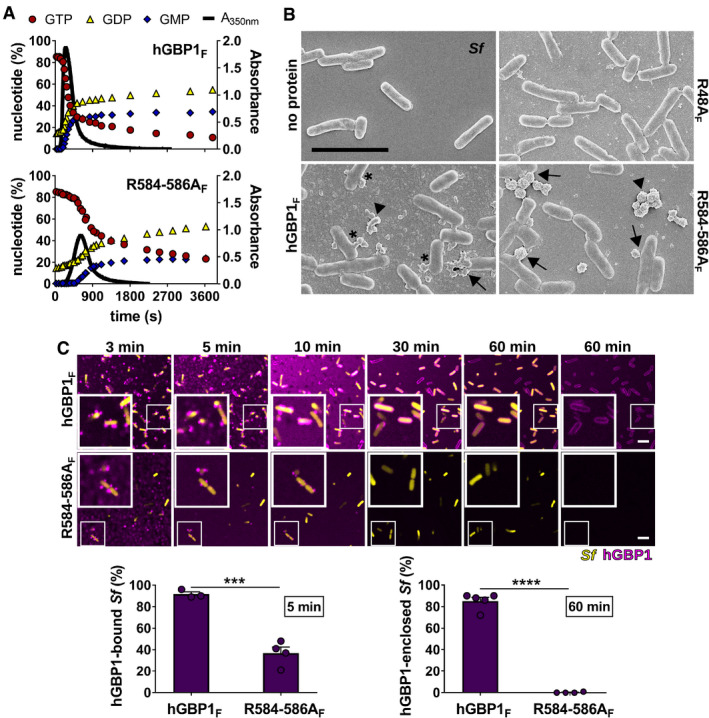
Figure 3. A C‐terminal hGBP1 polybasic motif is required for sustained binding to Shigella flexneri .
-
APolymerization of 10 μM hGBP1F and hGBP1F R584‐586A in the presence of 2 mM GTP was monitored over time by absorption spectroscopy at 350 nm. Absorbance signals were superimposed with nucleotide composition of the same solution analyzed at defined time points, revealing the characteristic first phase of slow polymer nucleation and the second phase of fast polymer growth and cooperative hydrolysis.
-
BScanning electron micrographs of live S. flexneri incubated with no protein or with 5 μM of either hGBP1F, hGBP1F R584‐586A, or hGBP1F R48A (non‐polymerizing mutant) in the presence of 2 mM GTP for 4 min. Arrowheads point to unattached hGBP1 polymers, arrows point to hGBP1 polymers attached to bacteria, and asterisks mark polymeric structures that appear to fuse with bacterial surfaces.
-
CConfocal time‐lapse microscopy frames of formaldehyde‐fixed GFP+ S. flexneri following admixture of 10 μM Alexa‐Fluor647‐labeled hGBP1F or hGBP1F R584‐586A and 2 mM GTP. Binding of hGBP1F polymers to bacteria at 5 min and enclosure of bacteria with hGBP1F protein coats after 60 min were quantified. Mean frequencies ± SEM of combined data from at least three independent experiments are shown. Significance was determined by unpaired t‐tests, two‐tailed. ***P ≤ 0.001; ****P ≤ 0.0001.
