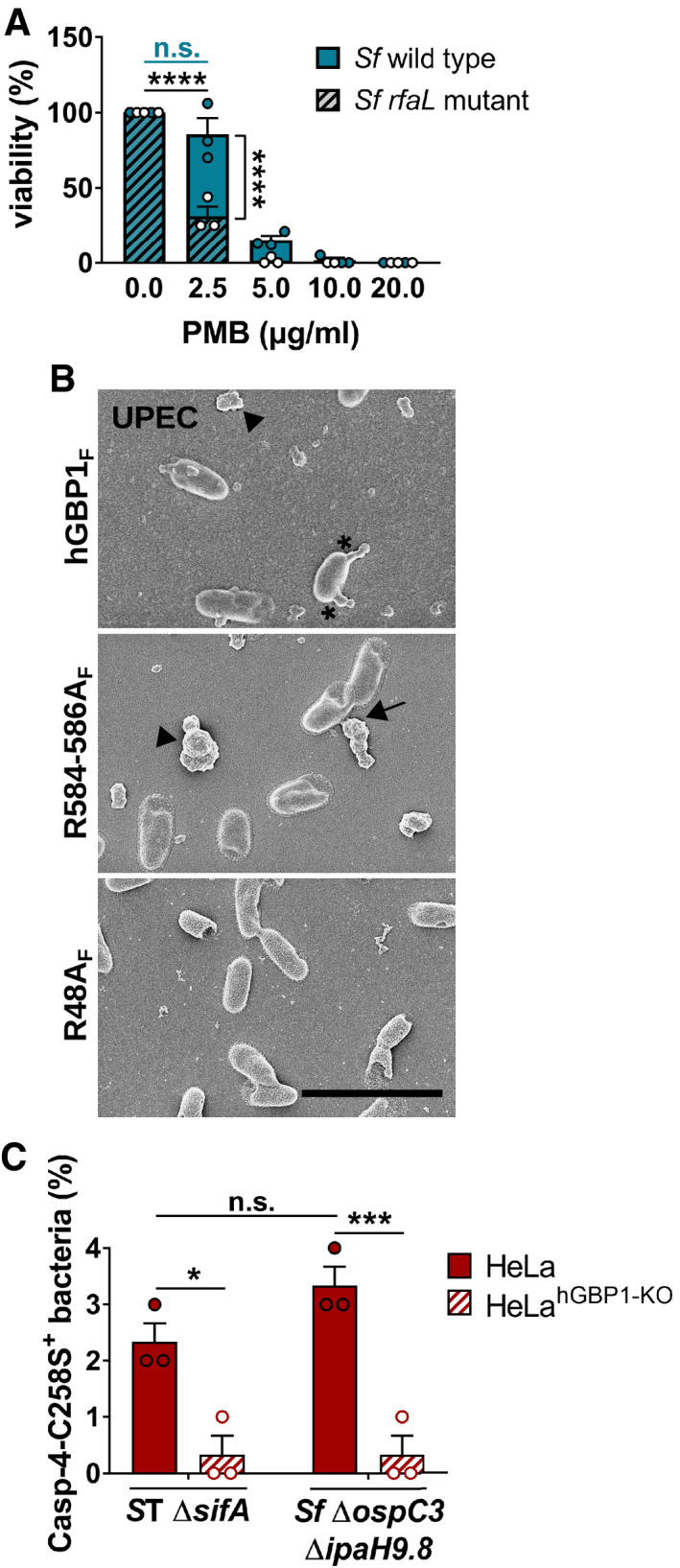
Figure EV4. Lack of O‐antigen renders Shigella flexneri more susceptible to polymyxin B.
-
AFollowing incubation for 45 min with buffer, live S. flexneri wild type and rfaL mutant were treated for 30 min with varying concentrations of polymyxin B (PMB). The number of viable bacteria was subsequently determined by CFU counts. Graphs show mean CFUs ± SEM of combined data from three independent experiments. Significance was determined by two‐way ANOVA with Tukey's multiple comparison test. n.s., not significant; ****P ≤ 0.0001.
-
BScanning electron micrographs of live UPEC incubated for 4 min with 5 μM hGBP1F or equivalent concentration of either hGBP1F R584‐586A or hGBP1F R48A in the presence of 2 mM GTP. Scale bar equals 5 μm. Arrowheads point to unattached hGBP1 polymers, arrows point to hGBP1 polymers attached to bacteria, and asterisks mark polymeric structures that appear to fuse with bacterial surfaces.
-
CIFNγ‐primed wild type and hGBP1‐KO HeLa cells stably expressing YFP‐Caspase‐4C258S were infected with either Salmonella enterica Typhimurium mutant ΔsifA (MOI = 25) or with S. flexneri ΔospC3ΔipaH9.8 (MOI = 6). Cells were fixed at 4 hpi (ST ΔsifA) or at 2 hpi (Sf ΔospC3ΔipaH9.8). Percentage of YFP‐Caspase‐4C258S‐associated bacteria were quantified, and mean ± SEM of combined data from three independent experiments are shown. Significance was determined by two‐way ANOVA with Tukey's multiple comparison test. n.s., not significant; *P ≤ 0.05; ***P ≤ 0.001.
Source data are available online for this figure.
