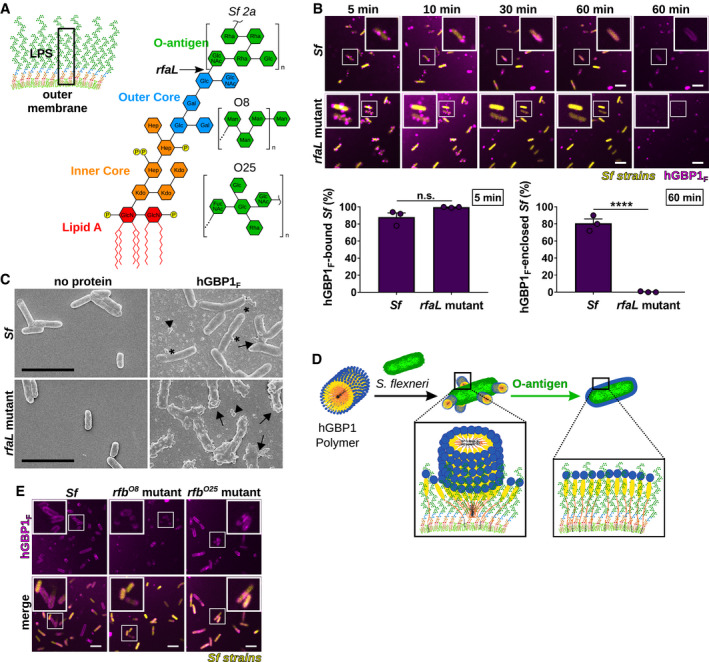
Figure 5. Bacterial O‐antigen drives the transition from bacteria‐bound hGBP1F polymers into bacteria‐encasing hGBP1F protein coats.
-
AGraphic depiction of the bacterial outer membrane composition and LPS structure of Shigella flexneri. Arrow indicates the LPS truncation site in the O‐antigen‐deficient rfaL mutant. O‐antigen oligosaccharide subunit composition of S. flexneri serotype 2a and E. coli serotypes O8 and O25 are shown.
-
BTime‐lapse microscopy of fixed GFP+ co‐isogenic S. flexneri wild type and rfaL mutant after adding 10 μM Alexa‐Fluor647‐hGBP1F and 2 mM GTP. Bacteria bound by hGBP1F at 5 min and hGBP1F‐enclosed bacteria after 60 min were quantified. Combined data from three independent experiments are shown as mean ± SEM. Significance was determined by unpaired t‐tests, two‐tailed. n.s., not significant; ****P ≤ 0.0001.
-
CScanning electron micrographs of live wild type and rfaL mutant strains incubated with no protein or with 5 μM of hGBP1F in the presence of 2 mM GTP for 4 min. Arrowheads point to unattached hGBP1 polymers, arrows point to hGBP1 polymers attached to bacteria, and asterisks mark polymeric structures that appear to fuse with bacterial surfaces.
-
DModel: O‐antigen drives the transition of surface‐docked hGBP1F polymers into bacteria‐enveloping hGBP1F protein sheets.
-
EConfocal images of formaldehyde‐fixed, co‐isogenic GFP+ wild type and mutant S. flexneri strains harboring rfb regions of E. coli serotypes O8 and O25 after 60 min of incubation time in the presence of 10 μM Alexa‐Fluor647‐hGBP1F and 2 mM GTP.
