Abstract
Light and temperature are two core environmental factors that coordinately regulate plant growth and survival throughout their entire life cycle. However, the mechanisms integrating light and temperature signaling pathways in plants remain poorly understood. Here, we report that CBF1, an AP2/ERF‐family transcription factor essential for plant cold acclimation, promotes hypocotyl growth under ambient temperatures in Arabidopsis. We show that CBF1 increases the protein abundance of PIF4 and PIF5, two phytochrome‐interacting bHLH‐family transcription factors that play pivotal roles in modulating plant growth and development, by directly binding to their promoters to induce their gene expression, and by inhibiting their interaction with phyB in the light. Moreover, our data demonstrate that CBF1 promotes PIF4/PIF5 protein accumulation and hypocotyl growth at both 22°C and 17°C, but not at 4°C, with a more prominent role at 17°C than at 22°C. Together, our study reveals that CBF1 integrates light and temperature control of hypocotyl growth by promoting PIF4 and PIF5 protein abundance in the light, thus providing insights into the integration mechanisms of light and temperature signaling pathways in plants.
Keywords: ambient temperature, CBF1, hypocotyl growth, phytochrome, PIF4/PIF5
Subject Categories: Plant Biology, Signal Transduction
Cold‐inducible transcription factor CBF1 integrates light and temperature control of seedling growth by inducing accumulation of light response proteins PIF4/PIF5.

Introduction
Plants have evolved sophisticated mechanisms to adapt to various environmental conditions. Light is a major environmental factor that regulates plant growth and development. Dark‐grown seedlings undergo skotomorphogenesis (etiolation), a developmental process characterized by long hypocotyls, closed and yellowish cotyledons, and apical hooks; by contrast, light‐grown seedlings undergo photomorphogenesis (de‐etiolation), a developmental process characterized by short hypocotyls, open and green cotyledons, and development of mature chloroplasts (Jiao et al, 2007; Li et al, 2011; Xu et al, 2015). Different wavelengths of light are perceived by distinct classes of photoreceptors, among which phytochromes are responsible for perceiving the red (R) and far‐red (FR) light spectrum (Jiao et al, 2007; Franklin & Quail, 2010; Li et al, 2011; Legris et al, 2019; Yadav et al, 2020). The Arabidopsis genome encodes five phytochrome proteins, named phyA to phyE. PhyA is the primary photoreceptor responsible for perceiving FR light, whereas phyB is the predominant photoreceptor regulating photomorphogenic responses to R light (Franklin & Quail, 2010; Li et al, 2011). Phytochromes exist in two interconvertible forms in vivo: the R‐absorbing Pr form and the FR‐absorbing Pfr form, and the Pfr form is generally considered to be the biologically active form (Bae & Choi, 2008; Li et al, 2011). In the dark, phytochromes are synthesized in the cytosol in the Pr form; upon exposure to R light, phytochromes are converted into the Pfr form, and translocate into the nucleus, where they rapidly change the expression of light‐responsive genes leading to the modulation of various biological responses (Jiao et al, 2007; Fankhauser & Chen, 2008; Franklin & Quail, 2010; Li et al, 2011; Legris et al, 2019).
Low temperature is a major abiotic stress that seriously affects the growth and development of plants. To survive cold stress, plants have evolved a suite of sophisticated strategies, among which cold acclimation is one of the major mechanisms plants use to adapt to low temperatures (Thomashow, 1999; Chinnusamy et al, 2007; Shi et al, 2015, 2018; Ding et al, 2019, 2020). Cold acclimation, a process by which plants acquire enhanced freezing tolerance upon prior exposure to low but nonlethal temperatures, is achieved by activation of a set of cold‐regulated (COR) genes, some of which encode the cryoprotective proteins or the key enzymes for osmolyte biosynthesis (Shi et al, 2018; Ding et al, 2019). Accumulating evidence indicates that three tandemly clustered C‐repeat binding factor/dehydration‐responsive element‐binding protein1 (CBF/DREB1) genes, encoding AP2/ERF‐family transcription factors, play central roles in plant cold acclimation. CBF proteins have been shown to directly bind CRT/DRE cis‐elements in the promoters of COR genes and activate their expression (Stockinger et al, 1997; Liu et al, 1998; Thomashow, 1999; Chinnusamy et al, 2007). Plants exposed to nonfreezing low temperatures were shown to induce CBF genes to high levels within 15 min (Gilmour et al, 1998; Medina et al, 1999; Vogel et al, 2005), thus leading to the activation of downstream COR genes and the enhancement of plant freezing tolerance. Emerging evidence has shown that CBF transcription factors are subjected to complicated but delicate control at transcriptional, post‐transcriptional, and post‐translational levels (Medina et al, 2011; Shi et al, 2015, 2018; Liu et al, 2018; Ding et al, 2019, 2020).
Phytochrome‐interacting factors (PIFs) are a group of basic helix‐loop‐helix (bHLH)‐family transcription factors. PIF3, the foundation member of the PIF subset, was firstly identified by a yeast two‐hybrid screen for phytochrome‐interacting factors (Ni et al, 1998). PIF4 was identified later by a forward genetic study, whereas PIF1 and PIF5 were characterized by their homology to PIF3 (Huq & Quail, 2002; Huq et al, 2004; Khanna et al, 2004; Oh et al, 2004). Later studies demonstrated that pif1 pif3 pif4 pif5 quadruple (abbreviated as pifq) mutants displayed a constitutively photomorphogenic (cop)‐like phenotype in darkness, demonstrating that the PIF proteins act to repress photomorphogenesis in etiolated seedlings (Leivar et al, 2008b; Shin et al, 2009). At the same time, compelling evidence indicated that upon illumination, photoactivated phytochromes interacted with nuclear PIF proteins to induce rapid phosphorylation, ubiquitination, and 26S proteasome‐mediated degradation of PIFs, thereby modulating the expression of a large number of PIF target genes within minutes (for reviews, see Leivar & Quail, 2011; Li et al, 2011; Leivar & Monte, 2014; Xu et al, 2015; Pham et al, 2018). Moreover, numerous studies have demonstrated that PIFs act as important integrators of diverse signaling pathways, including endogenous (e.g., hormonal) as well as external environmental cues (e.g., light and temperature), to optimize plant growth and development (Leivar & Monte, 2014; Paik et al, 2017).
As two key environmental factors, light and temperature signals are believed to be tightly associated, because in natural conditions light is normally accompanied by warm temperatures whereas dark by cooler temperatures. Consistent with this notion, it was shown that photoperiod and light quality regulated plant freezing tolerance through phytochromes (Kim et al, 2002; Franklin & Whitelam, 2007; Lee & Thomashow, 2012). The recent findings that the photoreceptor phyB functions as a thermosensor at ambient temperature (Jung et al, 2016; Legris et al, 2016), and that phototropin perceives both blue light and cold signals in the liverwort Marchantia polymorpha (Fujii et al, 2017) further reinforced this notion. HY5, a well‐characterized bZIP transcription factor playing a key role in promoting photomorphogenesis (Koornneef et al, 1980; Oyama et al, 1997), and COP1, an E3 ubiquitin ligase targeting HY5 for 26S proteasome‐mediated protein degradation (Osterlund et al, 2000), were shown to integrate cold and light signals to promote the cold acclimation response (Catalá et al, 2011; Noren et al, 2016). PIFs, including PIF3, PIF4, and PIF7, were shown to repress the expression of CBF genes under low temperatures by directly binding to their promoters, thus integrating light and temperature signals to optimize plant growth and cold tolerance (Kidokoro et al, 2009; Lee & Thomashow, 2012; Jiang et al, 2017). In addition, PIF4 has been shown to be a central regulator of warm‐temperature‐mediated elongation growth and flowering in Arabidopsis (Koini et al, 2009; Franklin et al, 2011; Kumar et al, 2012; Sun et al, 2012; Proveniers & van Zanten, 2013; Jung et al, 2016; Quint et al, 2016; Park et al, 2019; Vu et al, 2019). On the other hand, high expression of osmotically responsive genes 1 (HOS1), an E3 ubiquitin ligase originally identified as a cold signaling attenuator (Lee et al, 2001; Dong et al, 2006), was recently shown to participate in red light‐mediated degradation of constans (CO) (Lazaro et al, 2015) and facilitate phyB‐mediated inhibition of PIF4 function during hypocotyl growth (Kim et al, 2017).
In this paper, we report that CBF1 promotes hypocotyl growth under ambient temperatures in Arabidopsis. We show that CBF1 directly binds to the promoters of PIF4 and PIF5 and induces their expression in the light. In addition, our data demonstrate that CBF1 physically interacts with both phyA and phyB, and inhibits phyB interaction with PIF4 and PIF5. Moreover, our data demonstrate that CBF1 promotes hypocotyl growth and PIF4/PIF5 protein accumulation at 22°C and 17°C, but not at 4°C. Therefore, our study uncovers an important role of CBF1 in regulating Arabidopsis hypocotyl growth under ambient temperatures.
Results
CBF1 promotes hypocotyl growth in the light
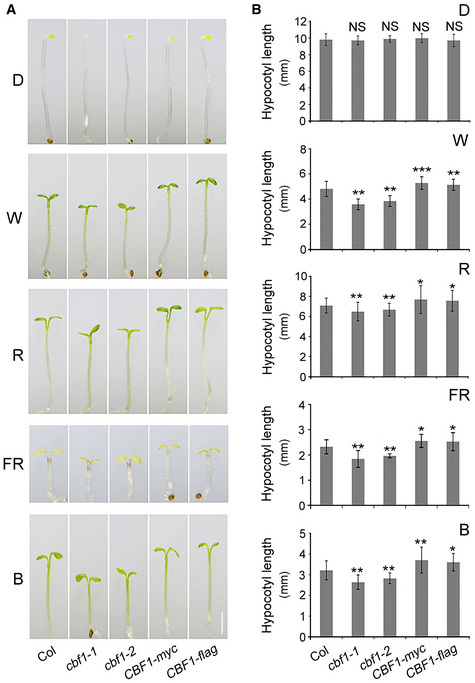
To investigate the mechanisms of cross‐talk between light and temperature signaling pathways, we asked whether CBFs, the pivotal transcription factors functioning in plant cold acclimation, could be involved in regulating Arabidopsis seedling photomorphogenesis. We first grew single mutant and overexpression (OE) lines of CBF1, CBF2, and CBF3 together with wild‐type (Col) in darkness (D) or continuous FR, R, blue (B), and white (W) light at 22°C for 4 days, and then compared their hypocotyl elongation and cotyledon expansion, two developmental processes regulated by light during the seedling stage (Jiao et al, 2007; Li et al, 2011; Sun et al, 2016). Interestingly, we observed that seedlings of two cbf1 mutants displayed shorter hypocotyls, whereas two CBF1‐OE lines exhibited longer hypocotyls in all tested light conditions but not in darkness (Fig 1). The short‐hypocotyl phenotype of cbf1 mutants in the light was not due to delayed seed germination (Appendix Fig S1A). In addition, the cotyledon areas of CBF1‐OE lines were significantly larger than those of Col, whereas cbf1 mutants did not display any detectable changes in cotyledon areas (Appendix Fig S1B and C). Together, our data indicated that CBF1 promoted the growth of hypocotyls in the light, while its role in regulating cotyledon expansion needs to be further characterized.
Figure 1. CBF1 promotes Arabidopsis hypocotyl elongation in the light.
-
APhenotypes of 4‐day‐old Col, two cbf1 mutants (cbf1‐1 and cbf1‐2), and two CBF1‐OE lines (CBF1‐myc and CBF1‐flag) grown at 22°C in darkness (D) or continuous W, R, FR, and B light. Scale bar = 1 mm.
-
BHypocotyl lengths of Col, two cbf1 mutants, and two CBF1‐OE lines grown at 22°C in D or continuous W, R, FR, and B light. Error bars represent SD from 20 seedlings. *P < 0.05, **P < 0.01, and ***P < 0.001 (two‐tailed t‐test) for the indicated genotype compared with Col. NS, not significant.
In addition, we found that lines over‐expressing CBF2 or CBF3 showed significantly shorter hypocotyls in darkness and all tested light conditions (Appendix Fig S2). We reasoned that these phenotypes may be caused by growth retardation due to overexpression of CBFs, which was reported previously (Jaglo‐Ottosen et al, 1998; Kasuga et al, 1999; Gilmour et al, 2000, 2004; Achard et al, 2008). The monogenic cbf2 and cbf3 mutants did not display any detectable photomorphogenic phenotypes except that the cbf2 mutants developed longer hypocotyls under continuous blue light (Appendix Fig S2). Therefore, we selected CBF1 for further investigation. These observations also suggested that the three CBF proteins may play distinct roles during plant growth and development.
The transcript and protein levels of CBF1 are induced by light
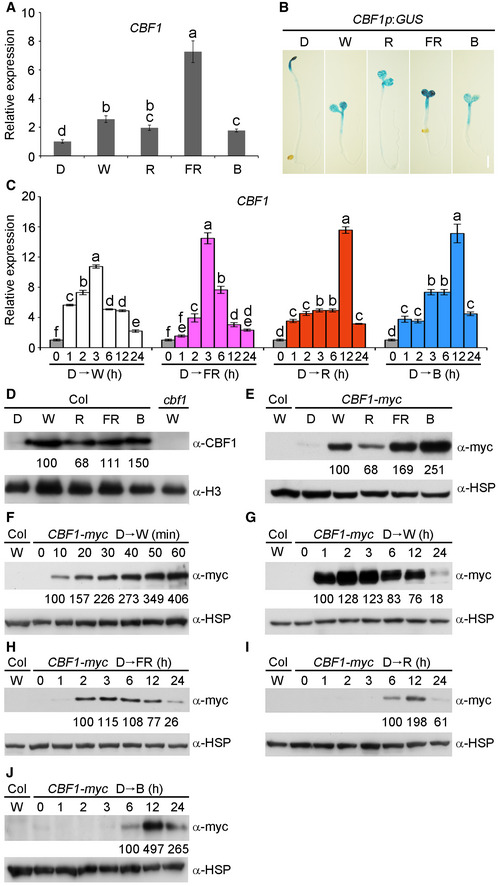
We then investigated whether CBF1 expression was regulated by light. We first examined CBF1 transcript levels in 4‐day‐old wild‐type (Col) seedlings grown at 22°C in darkness or continuous FR, R, B, and W light by qRT–PCR. Our data indicated that CBF1 expression was elevated in light‐grown relative to dark‐grown seedlings, especially in FR light under which CBF1 displayed the highest expression (Fig 2A). Interestingly, our qRT–PCR data showed that CBF2 and CBF3 expression was also regulated by light but in different patterns (Appendix Fig S3).
Figure 2. CBF1 transcript and protein levels are induced by light.
-
AqRT–PCR analysis showing the relative expression of CBF1 in 4‐day‐old wild‐type (Col) seedlings grown at 22°C in darkness (D) or continuous W, R, FR, and B light. Error bars represent SD of three technical replicates. Different letters represent significant differences by one‐way ANOVA with Duncan's post hoc test (P < 0.05).
-
BGUS staining of 4‐day‐old homozygous CBF1p:GUS transgenic seedlings grown at 22°C in D or continuous W, R, FR, and B light. Scale bar = 1 mm.
-
CqRT–PCR data showing that CBF1 expression is induced by different light regimes. Wild‐type (Col) seedlings were first grown at 22°C in D for 4 days and then transferred to continuous W, FR, R, or B light for the indicated times ranging from 1 to 24 h. Error bars represent SD of three technical replicates. Different letters represent significant differences by one‐way ANOVA with Duncan's post hoc test (P < 0.05).
-
DImmunoblots showing CBF1 protein levels in nuclear fractions from 4‐day‐old wild‐type (Col) seedlings grown at 22°C in D or continuous W, R, FR, and B light.
-
EImmunoblots showing CBF1‐myc protein levels in 4‐day‐old CBF1‐myc seedlings grown at 22°C in D or continuous W, R, FR, and B light.
-
F, GImmunoblots showing the levels of CBF1‐myc proteins after exposure to white light. CBF1‐myc seedlings were first grown at 22°C for 4 days in D and then transferred to continuous W light for the indicated time periods within 1 h (F) or ranging from 1 to 24 h (G).
-
H–JImmunoblots showing the levels of CBF1‐myc proteins after exposure to FR (H), R (I) or B (J) light. CBF1‐myc seedlings were first grown at 22°C for 4 days in D and then transferred to continuous FR, R, or B light for the indicated time periods ranging from 1 to 24 h.
To understand whether light changes the spatial expression of CBF1, we generated CBF1p:GUS lines in which GUS gene expression was driven by the native CBF1 promoter. The histochemical staining results indicated that GUS was mainly expressed in cotyledons and upper hypocotyls in both light and dark (Fig 2B). Consistent with our qRT–PCR data, CBF1 was highly expressed in the cotyledons under FR light (Fig 2A and B). To further confirm the light‐regulated expression of CBF1, we transferred 4‐day‐old dark‐grown wild‐type (Col) seedlings to continuous FR, R, B, and W light for 1–24 h. The materials were then harvested and subjected to qRT–PCR assays. Our data showed that, indeed, the CBF1 transcript level was dramatically induced (up to 10‐ to 20‐fold) under different light conditions (Fig 2C). However, CBF1 expression was rapidly induced by FR and W light (peaking at 3 h after transfer), whereas it was induced relatively more slowly by R and B light (peaking at 12 h after transfer; Fig 2C). Together, our data demonstrated that CBF1 expression was induced by light.
Next, we asked whether CBF1 protein accumulation was also regulated by light. We generated polyclonal anti‐CBF1 antibodies, but endogenous CBF1 proteins were hardly detectable from total proteins of dark‐ or light‐grown wild‐type seedlings, possibly due to the extremely low levels of CBF1 proteins in vivo. However, we were able to detect endogenous CBF1 proteins in the nuclear fractions prepared from wild‐type (Col) seedlings grown under continuous light using our anti‐CBF1 antibodies (Fig 2D). In addition, our immunoblots of total proteins extracted from 35S:CBF1‐myc (abbreviated as CBF1‐myc) seedlings showed that the pattern of CBF1‐myc protein accumulation was similar to that of nuclear CBF1 under the tested conditions, i.e., CBF1 accumulated in the light but not in the dark (Fig 2D and E). Therefore, we used CBF1‐myc seedlings to further investigate how light regulated the abundance of CBF1 proteins. Strikingly, after we transferred 4‐day‐old dark‐grown CBF1‐myc seedlings to continuous W light, we detected CBF1 within 10 min after transfer (Fig 2F). CBF1 proteins continued to accumulate in W light, peaking at 2–3 h after transfer and then gradually decreased after prolonged exposure to a low level at 24 h (Fig 2G). Similar regulation of CBF1 was observed after CBF1‐myc seedlings were transferred to FR light (Fig 2H). However, delayed induction of CBF1 was observed after exposure to R and B light, under which CBF1 protein levels both peaked at 12 h after transfer (Fig 2I and J). These data indicated that CBF1 protein levels were differentially controlled in different light regimes. Notably, the induction patterns of CBF1 proteins were similar to those of CBF1 transcripts upon exposure to W, FR, R, and B light, respectively (Fig 2C). Collectively, our data demonstrated that both CBF1 transcript and protein levels were induced by light, but displayed distinct induction patterns under varying light conditions.
CBF1 transcript and protein levels are induced by phyA and phyB in R light
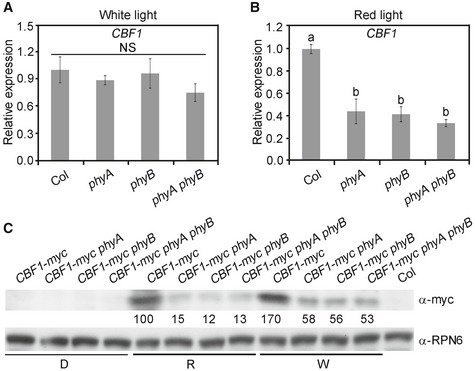
Due to the pivotal role of phytochromes in perceiving light signals in plants, we next examined how CBF1 expression was regulated by phyA and phyB. We grew wild‐type (Col), phyA, phyB, and phyA phyB mutant seedlings in continuous W and R light at 22°C for 4 days, and then examined the CBF1 transcript levels by qRT–PCR. Our data showed that phyA and phyB played a minor role in regulating CBF1 expression in W light; by contrast, CBF1 transcript levels were drastically decreased in phyA, phyB, and phyA phyB mutants in R light (Fig EV1A and B), indicating that phyA and phyB mediate R light induction of CBF1 expression.
Figure EV1. CBF1 transcript and protein levels are induced by phyA and phyB in red light.
-
A, BqRT–PCR assays showing the expression of CBF1 in 4‐day‐old Col, phyA, phyB, and phyA phyB seedlings grown at 22°C in continuous W (A) or R light (B). Error bars represent SD of three technical replicates. Different letters represent significant differences by one‐way ANOVA with Duncan's post hoc test (P < 0.05). NS, not significant.
-
CImmunoblots showing the levels of CBF1‐myc proteins in 4‐day‐old CBF1‐myc, CBF1‐myc phyA, CBF1‐myc phyB, and CBF1‐myc phyA phyB seedlings grown at 22°C in darkness (D) or continuous R or W light. Anti‐RPN6 was used as a sample loading control. Numbers below the immunoblot indicate the relative band intensities of CBF1‐myc normalized to those of loading control, respectively. The ratio of the first clear band was set to 100.
Source data are available online for this figure.
We then examined how phyA and phyB regulated the accumulation of CBF1 proteins in the light. To this end, we introduced phyA, phyB, and phyA phyB mutations into 35S:CBF1‐myc, respectively, by genetic crossing, and obtained lines homozygous for the respective loci (Appendix Fig S4). We then grew CBF1‐myc, CBF1‐myc phyA, CBF1‐myc phyB, and CBF1‐myc phyA phyB seedlings in the dark or continuous R or W light for 4 days. Our immunoblot data indicated that the levels of 35S‐driven CBF1‐myc decreased in the absence of phyA or phyB under these two light conditions, especially in R light (Fig EV1C), suggesting that phyA and phyB were involved in post‐translational regulation of CBF1 protein stability in R and W light. Together, our data demonstrated that both CBF1 transcript and protein levels were induced by phyA and phyB in R light.
Genetic relationship between CBF1 and phyA/phyB
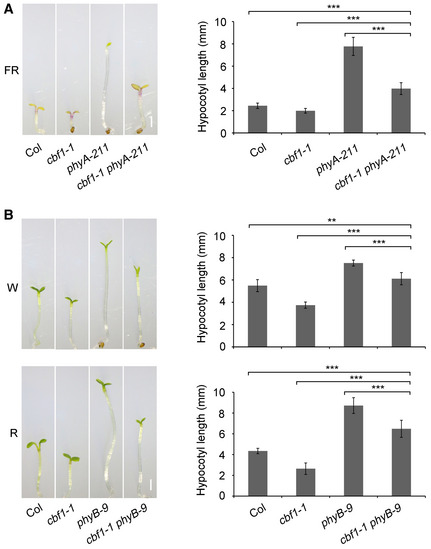
To investigate the genetic relationships between CBF1 and phyA/phyB, we generated cbf1‐1 phyA‐211 and cbf1‐1 phyB‐9 double mutants by crossing cbf1‐1 with phyA‐211 and phyB‐9, respectively. Genotyping data showed that the corresponding mutated loci were homozygous in the respective double mutants (Appendix Fig S5). The cbf1‐1 phyA‐211 mutants were grown at 22°C in continuous FR light together with Col, phyA‐211, and cbf1‐1 for 4 days, and we observed that the hypocotyl lengths of cbf1‐1 phyA‐211 mutant seedlings were shorter than those of phyA, but longer than those of Col seedlings (Fig EV2A). The cbf1‐1 phyB‐9 mutants were then grown at 22°C in continuous W and R light for 4 days, and we also observed that cbf1‐1 phyB‐9 mutant seedlings also developed intermediate hypocotyl lengths compared with Col and phyB mutant seedlings in both W and R light (Fig EV2B). These observations indicated that CBF1 was indispensable for the long‐hypocotyl phenotypes of phyA‐211 mutant seedlings in FR light and of phyB‐9 mutant seedlings in W and R light, respectively.
Figure EV2. Genetic relationship between CBF1 and phyA/phyB.
-
APhenotypes and hypocotyl lengths of 4‐day‐old Col, cbf1‐1, phyA‐211, and cbf1‐1 phyA‐211 seedlings grown at 22°C in continuous FR light.
-
BPhenotypes and hypocotyl lengths of 4‐day‐old Col, cbf1‐1, phyB‐9, and cbf1‐1 phyB‐9 seedlings grown at 22°C in continuous W or R light.
CBF1 interacts with phyA and phyB
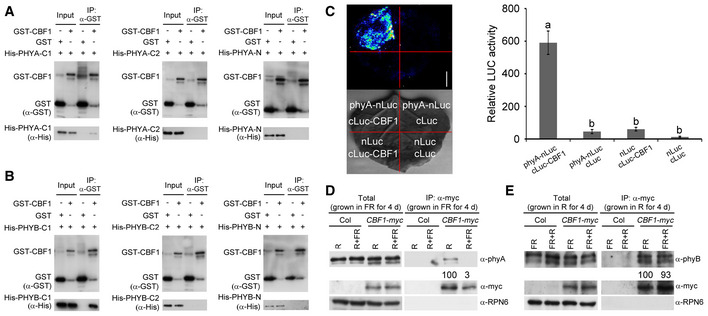
Next, we asked whether CBF1 could physically interact with phyA and phyB. We first performed in vitro pull‐down assays to address this question. His‐tagged N‐terminal chromophore‐bearing photosensory domains (designated N), PAS‐related domains (PRD, designated C1) and histidine kinase‐related domains (HKRD, designated C2) of PHYA or PHYB (Appendix Fig S6), and GST‐tagged CBF1 (designated as GST‐CBF1) were expressed and purified from Escherichia coli. Our pull‐down assays showed that GST‐tagged CBF1, but not GST alone, was able to pull down His‐tagged C1 domains of both PHYA and PHYB in vitro (Fig 3A and B). His‐tagged N or C2 domains of both PHYA and PHYB were not pulled down by GST‐tagged CBF1 (Fig 3A and B). A parallel assay also showed that GST‐tagged C1 domains of both PHYA and PHYB, but not other domains of PHYA or PHYB, or GST alone, were able to pull down His‐CBF1 in vitro (Appendix Fig S7). These data indicated that CBF1 could interact with the PAS‐related domains of both PHYA and PHYB in vitro, and this interaction was not affected by different fusion tags.
Figure 3. CBF1 physically interacts with phyA and phyB.
-
A, BIn vitro pull‐down of PHYA‐C1 (A) and PHYB‐C1 (B) with CBF1. The His‐tagged PHYA/B‐N (photosensory domain), PHYA/B‐C1 (PAS‐related domain), and PHYA/B‐C2 (histidine kinase‐related domain) proteins pulled down with GST‐tagged CBF1 or GST were detected by anti‐His antibody. Input, 6% of the His‐tagged purified target proteins used in pull‐down assays.
-
CLCI assays showing that CBF1 interacted with phyA in living plant cells. Representative picture is shown in the left panel, and relative luciferase activity is shown in the right panel. Data are the means ± SD of three independent biological repeats. Different letters represent significant differences by one‐way ANOVA with Duncan's post hoc test (P < 0.05). Scale bar = 1 cm.
-
D, ECo‐IP assays showing that CBF1 interacted with phyA (D) and phyB (E) in vivo. (D) Homozygous CBF1‐myc seedlings were first grown at 22°C in continuous FR light for 4 days, and then, the total proteins were extracted and subsequently treated with 5‐min R light or 5‐min R light followed by 5‐min FR light (R + FR) before immunoprecipitation. (E) Homozygous CBF1‐myc seedlings were first grown at 22°C in continuous R light for 4 days, and then, the total proteins were extracted and subsequently treated with 5‐min FR light or 5‐min FR light followed by 5‐min R light (FR + R) before immunoprecipitation. After light treatments, total proteins were incubated with anti‐c‐myc Affinity Gel (Sigma‐Aldrich). The total and precipitated proteins were examined by immunoblotting using antibodies against myc, phyA, phyB, and RPN6, respectively. The numbers below anti‐phyA and anti‐phyB blots indicate the relative band intensities of co‐precipitated phyA or phyB normalized to those of precipitated CBF1‐myc, respectively. The ratio of the first clear band was set to 100 for each blot.
Source data are available online for this figure.
To verify the physical interaction between CBF1 and phyA in planta, firefly luciferase complementation imaging (LCI) assays (Chen et al, 2008) were performed by transiently expressing phyA‐nLuc and cLuc‐CBF1 fusions in Nicotiana benthamiana leaf cells. Our data showed that co‐expression of phyA‐nLuc and cLuc‐CBF1 led to strong LUC activity, whereas phyA‐nLuc or cLuc‐CBF1 co‐transformed with control vectors showed only background levels of LUC activity (Fig 3C). These observations support the conclusion that CBF1 physically interacts with phyA in living plant cells. To confirm the physical interaction between CBF1 and phyA in vivo, we conducted co‐immunoprecipitation (co‐IP) assays using 4‐day‐old CBF1‐myc seedlings grown at 22°C in continuous FR light. To determine which form (Pr or Pfr) of phyA associated with CBF1 more strongly, total proteins were exposed to 5 min of R light (for conversion of phytochromes to the Pfr form) or 5 min of R light immediately followed by 5 min of FR light (for conversion back to the Pr form). Our immunoblot data showed that phyA was co‐precipitated with anti‐myc antibodies in CBF1‐myc, but not in Col seedlings (Fig 3D). Moreover, phyA was apparently co‐precipitated with CBF1‐myc only after R light exposure (Fig 3D), indicating that CBF1 preferentially interacted with the Pfr form of phyA in vivo. We also grew the CBF1‐myc seedlings at 22°C in continuous R for 4 days and then conducted light pulse treatments followed by co‐IP assays to examine the in vivo association of CBF1 with phyB. Our results showed that similar amounts of phyB were co‐precipitated with CBF1‐myc after FR or FR + R light pulse treatments (Fig 3E), indicating that CBF1 interacted with the Pr and Pfr forms of phyB with similar affinity in vivo. Together, our data indicated that CBF1 interacted with both phyA and phyB in vivo, but more selectively with phyA than with phyB.
CBF1 positively regulates PIF4 and PIF5 expression by directly binding to their promoters
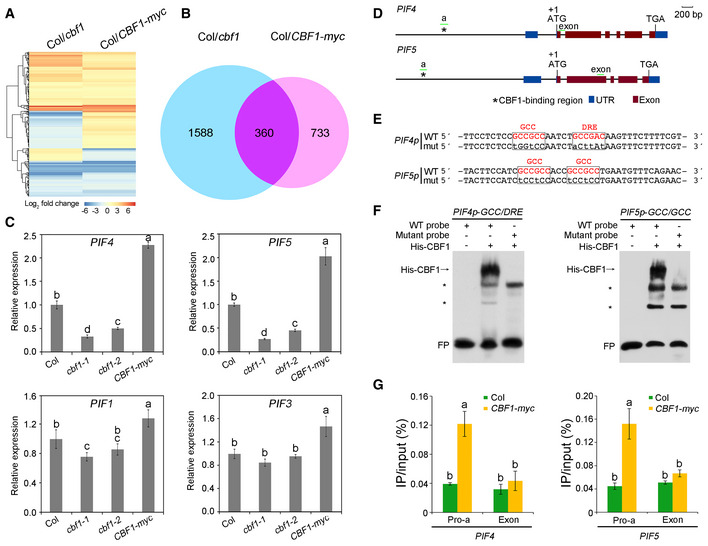
To explore the potential genes CBF1 targets to regulate light signaling, we examined the transcriptomes of 4‐day‐old Col, cbf1‐1, and CBF1‐myc seedlings grown at 22°C under continuous W light by RNA‐sequencing (RNA‐seq) analysis. The RNA‐seq data were collected from three independent biological experiments (each sample with 2.0 G clean data) and differential gene expression analysis was performed using Cufflinks (Trapnell et al, 2013) (cufflinks.cbcb.umd.edu). Our data revealed 1,948 and 1,093 genes, respectively, in cbf1‐1 and CBF1‐myc seedlings whose expression displayed statistically significant changes (using Student's t‐test with P < 0.05 and fold change > 2; Fig EV3A and B). Further analyses showed that a total of 360 genes exhibited at least twofold expression changes in both cbf1‐1 and CBF1‐myc seedlings (Fig EV3B). Among these, PIF4 and PIF5, two key regulatory genes of the light signaling pathway, were shown to be significantly regulated by CBF1 (Dataset EV1). However, several other key regulatory genes of the phytochrome signaling pathway, including PHYA, PHYB, PIF1, PIF3, COP1, and HY5, did not display twofold expression changes in RNA‐seq in either cbf1‐1 or CBF1‐myc seedlings. We then performed qRT–PCR assays to further validate the RNA‐seq results, and our data indicated that, indeed, the expression of PIF4 and PIF5 was significantly induced by CBF1 (Fig EV3C). In addition, COP1 was shown to be slightly induced by CBF1, whereas PIF1, PIF3, PHYA, and PHYB were not evidently regulated by CBF1 in qRT–PCR assays (Fig EV3C, Appendix Fig S8). Notably, our qRT–PCR data showed that CBF1 regulation of PIF4 and PIF5 expression predominates in the light relative to the dark (Fig EV3C, Appendix Fig S9).
Figure EV3. CBF1 positively regulates PIF4 and PIF5 expression by directly binding to their promoters.
-
ACluster analysis of genes whose expression was changed in cbf1 and CBF1‐myc seedlings compared with Col. The bar represents the log2 of the ratio.
-
BVenn diagram showing the numbers and overlaps of genes whose expression was changed in cbf1 and CBF1‐myc seedlings.
-
CqRT–PCR assays showing the expression levels of PIF4, PIF5, PIF1, and PIF3 in 4‐day‐old Col, cbf1‐1, cbf1‐2, and CBF1‐myc seedlings grown at 22°C in continuous W light. Error bars represent SD of three technical replicates. Different letters represent significant differences by one‐way ANOVA with Duncan's post hoc test (P < 0.05).
-
DSchematic illustration of the exon–intron structures of PIF4 and PIF5. Asterisks indicate the CBF1‐binding regions in the PIF4 and PIF5 promoters, and the sequences are shown in (E). The short green lines depict the location of amplicons used for ChIP‐qPCR shown in (G). UTR, untranslated region.
-
EWild‐type (WT) and mutated (mut) EMSA probes according to the sequences of CBF1‐binding regions in the PIF4 and PIF5 promoters. Wild‐type GCC or DRE elements are shown in red, and the nucleotide substitutions in the mutant fragments are shown as lowercase letters.
-
FEMSAs showing that His‐CBF1 directly binds the wild‐type, but not mutant probes of the PIF4 and PIF5 promoter fragments. Asterisks indicate nonspecific binding. FP, free probe.
-
GChIP assays showing that CBF1 directly binds to PIF4 and PIF5 promoters in vivo. Four‐day‐old Col and CBF1‐myc seedlings grown at 22°C in continuous white light were harvested and subjected to ChIP analysis using anti‐c‐myc Affinity Gel (Sigma‐Aldrich), and the precipitated DNA was recovered and analyzed by qPCR assays. The locations of amplicons used for ChIP‐qPCR assays are shown in (D). Error bars represent SD of three technical replicates. Different letters represent significant differences by one‐way ANOVA with Duncan's post hoc test (P < 0.05).
Source data are available online for this figure.
It was interesting to notice that CBF1 could positively regulate PIF4 and PIF5 expression in the light. We then asked whether CBF1 could directly bind to their promoters. Previous reports showed that several AP2/ERF‐family transcription factors could directly bind the cis‐elements, CRT/DRE (RCCGAC) and GCC (GCCGCC) (Stockinger et al, 1997; Gilmour et al, 1998; Liu et al, 1998; Zhu et al, 2014; Shi et al, 2018). Further analyses revealed the presence of a short sequence (~ 20 bp) in both PIF4 and PIF5 promoters which contained two consecutive motifs of either CRT/DRE or GCC (Fig EV3D and E). To investigate whether CBF1 could directly bind to these motifs, we performed electrophoretic mobility shift assays (EMSAs) using His‐tagged CBF1 proteins and biotin‐labeled wild‐type (WT) or mutant probes. Our EMSA data showed that CBF1 was able to bind the WT probes of both PIF4 and PIF5 promoters in vitro, whereas mutations of the two consecutive motifs abolished CBF1 binding to these probes (Fig EV3F), indicating that CBF1 bound to the CRT/DRE and GCC motifs present in the PIF4 and PIF5 promoters.
To confirm that CBF1 bound the PIF4 and PIF5 promoters in vivo, we performed chromatin immunoprecipitation (ChIP) assays using 4‐day‐old Col and CBF1‐myc seedlings grown at 22°C in continuous white light. Our qPCR data indicated that the amplicons of PIF4 and PIF5 promoters, which included the CRT/DRE or GCC motifs, were highly enriched in the anti‐myc ChIP samples of CBF1‐myc, but not in those of Col seedlings, as compared with the enrichment of exonic regions of PIF4 and PIF5 (Fig EV3G). These results indicated that CBF1 bound to these PIF4 and PIF5 promoter fragments in vivo. Collectively, our data demonstrated that CBF1 positively regulated PIF4 and PIF5 expression by directly binding to their promoters.
CBF1 promotes PIF4 and PIF5 protein accumulation in the light
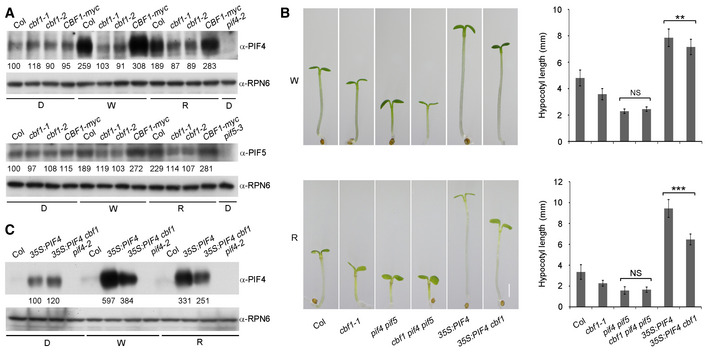
Next, we asked whether CBF1 regulated PIF4 and PIF5 protein accumulation under different light regimes. We grew Col, two cbf1 mutants (cbf1‐1 and cbf1‐2), and CBF1‐myc seedlings in darkness or continuous W or R light at 22°C for 4 days, and then examined the levels of PIF4 and PIF5 proteins by immunoblotting. Our data showed that PIF4 and PIF5 protein levels were much lower in the cbf1 mutants in both continuous W and R light (Fig 4A), indicating that CBF1 positively regulated PIF4 and PIF5 protein accumulation in the light. By contrast, CBF1 did not appear to regulate PIF4 and PIF5 protein accumulation in darkness (Fig 4A). These observations were consistent with our qRT–PCR data that CBF1 regulates PIF4 and PIF5 expression mainly in the light (Fig EV3C, Appendix Fig S9).
Figure 4. CBF1 promotes PIF4 and PIF5 protein accumulation in the light.
-
AImmunoblots showing the levels of PIF4 and PIF5 proteins at 22°C in 4‐day‐old Col, cbf1‐1, cbf1‐2, and CBF1‐myc seedlings grown in darkness (D) or continuous W or R light. Anti‐RPN6 was used as a sample loading control.
-
BPhenotypes and hypocotyl lengths of 4‐day‐old Col, cbf1‐1, pif4 pif5, cbf1 pif4 pif5, 35S:PIF4, and 35S:PIF4 cbf1 seedlings grown at 22°C in continuous W or R light. Error bars represent SD from 20 seedlings. **P < 0.01 and ***P < 0.001 (two‐tailed t‐test) for the indicated pairs of seedlings. Scale bar = 1 mm. NS, not significant.
-
CImmunoblots showing the PIF4 protein levels in 4‐day‐old Col, 35S:PIF4, and 35S:PIF4 cbf1 seedlings grown at 22°C in D or continuous W or R light. Anti‐RPN6 was used as a sample loading control.
To further investigate whether CBF1 promotes hypocotyl elongation in the light by positively regulating PIF4 and PIF5, we crossed cbf1‐1 with pif4 pif5 and obtained homozygous cbf1 pif4 pif5 triple mutants (Appendix Fig S10A–C). The cbf1 pif4 pif5 mutants were then grown together with Col, cbf1‐1, and pif4 pif5 mutants in continuous W and R light at 22°C for 4 days. We observed that the hypocotyl lengths of cbf1 pif4 pif5 mutants were indistinguishable from those of pif4 pif5 in both W and R light conditions (Fig 4B). These observations suggest that pif4 pif5 mutations are epistatic to cbf1 in regulating hypocotyl elongation in the light, consistent with the conclusion that PIF4 and PIF5 were downstream target genes of CBF1.
We also crossed 35S:PIF4 into the cbf1‐1 background (Appendix Fig S10D) and grew 35S:PIF4 cbf1‐1 and 35S:PIF4 seedlings in continuous W and R light at 22°C for 4 days. Interestingly, we found that 35S:PIF4 cbf1 developed significantly shorter hypocotyls than 35S:PIF4 seedlings in continuous W and R light (Fig 4B). Consistent with this observation, whereas PIF4 levels were largely similar in dark‐grown 35S:PIF4 cbf1 and 35S:PIF4 seedlings, PIF4 proteins accumulated to lower levels in 35S:PIF4 cbf1 than in 35S:PIF4 seedlings in continuous W and R light (Fig 4C), suggesting that CBF1 promotes PIF4 protein abundance in the light in ways additional to transcriptional upregulation of PIF4 expression.
CBF1 inhibits phyB interaction with PIF4 and PIF5
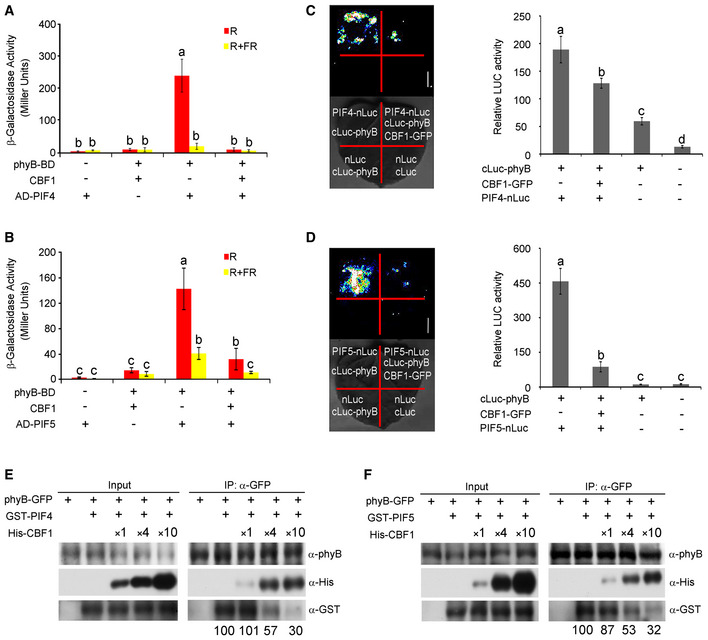
The fact that 35S‐driven PIF4 accumulated to lower levels in the light in the absence of CBF1 led us to speculate that CBF1 may promote PIF4 protein accumulation through post‐translational mechanisms as well. It was previously documented that phytochrome interaction with PIFs induced their rapid turnover upon light exposure (Bauer et al, 2004; Al‐Sady et al, 2006; Shen et al, 2007, 2008; Bae & Choi, 2008; Lorrain et al, 2008; Leivar & Quail, 2011; Ni et al, 2013). We therefore asked whether CBF1 might regulate phyB interaction with PIF4 and PIF5. We first performed yeast three‐hybrid assays by adding the chromophore phycocyanobilin (PCB) extracted from Spirulina to allow the phytochromes to form Pr or Pfr forms in yeast cells after FR or R light pulse treatments, respectively. Consistent with previous reports that PIF4 and PIF5 interacted with the Pfr form of phyB (Huq & Quail, 2002; Khanna et al, 2004), our data indicated that AD‐PIF4 and AD‐PIF5 interacted with phyB‐BD in yeast cells predominantly after R light pulse treatments (Fig 5A and B). Interestingly, when CBF1 was co‐expressed in yeast cells, the interactions between AD‐PIF4/PIF5 and the Pfr form of phyB‐BD were dramatically decreased (Fig 5A and B). Immunoblot data excluded the possibility that the decreased interactions were due to lower levels of AD‐PIF4/PIF5 or phyB‐BD proteins caused by co‐expression of CBF1 in yeast cells (Appendix Fig S11). These data demonstrated that CBF1 inhibited the interactions between PIF4/PIF5 and the Pfr form of phyB in yeast cells.
Figure 5. CBF1 inhibits phyB interaction with PIF4 and PIF5.
-
A, BYeast three‐hybrid assays showing that CBF1 represses the interaction of the Pfr‐phyB with PIF4 (A) and PIF5 (B) in yeast cells. AD‐PIF4, AD‐PIF5, phyB‐BD, and CBF1 were expressed in the yeast strain Y190 as indicated. The β‐galactosidase activities were measured by liquid culture assays using ONPG as the substrate. Error bars represent SD of three independent yeast clones. Different letters represent significant differences by one‐way ANOVA with Duncan's post hoc test (P < 0.05).
-
C, DLCI assays showing that CBF1 inhibits phyB interaction with PIF4 (C) and PIF5 (D) in plant cells. Representative picture is shown in the left panel, and relative luciferase activity is shown in the right panel. Data are the means ± SD of three independent biological repeats. Different letters represent significant differences by one‐way ANOVA with Duncan's post hoc test (P < 0.05).
-
E, FSemi‐in vivo pull‐down assays showing that CBF1 inhibits phyB interaction with PIF4 (E) and PIF5 (F). The total proteins extracted from 4‐day‐old 35S:phyB‐GFP seedlings grown at 22°C in continuous R light served as the bait, and GST‐PIF4 or GST‐PIF5 fusion proteins served as the prey. Equivalent amounts of phyB‐GFP protein extract and GST‐PIF4 or GST‐PIF5 fusion proteins, and increasing amounts of His‐CBF1 were added as indicated before anti‐GFP IP assays were performed. The pulled‐down proteins were subjected to immunoblotting with antibodies against phyB, GST, and His, respectively. Numbers below the anti‐GST blots indicate the relative band intensities of co‐precipitated GST‐PIF4 or GST‐PIF5 normalized to those of precipitated phyB‐GFP, respectively. The ratio of the first clear band was set to 100 for each blot.
Source data are available online for this figure.
To further confirm that CBF1 acts to disrupt the interactions between phyB and PIF4/PIF5 in planta, we performed LCI assays to detect interactions between PIF4‐nLuc or PIF5‐nLuc with cLuc‐phyB in tobacco leaves in the presence and absence of CBF1. Our LCI data showed that both PIF4‐nLuc and PIF5‐nLuc interacted strongly with cLuc‐phyB, respectively, in tobacco leaves in the absence of CBF1; in contrast, co‐expression of CBF1 led to a drastic decrease in the interactions of both PIF4‐nLuc and PIF5‐nLuc with cLuc‐phyB (Fig 5C and D). We also performed semi‐in vivo pull‐down assays using GST‐PIF4/PIF5 and His‐CBF1 fusion proteins expressed in E. coli, and total proteins extracted from 4‐day‐old phyB‐GFP seedlings grown at 22°C in continuous R light. We first incubated GST‐PIF4 or GST‐PIF5 with the protein extracts prepared from phyB‐GFP seedlings and then performed anti‐GFP IP assays. Our immunoblot data showed that GST‐PIF4 and GST‐PIF5 were indeed co‐precipitated with phyB‐GFP (Fig 5E and F). However, when we added increasing amounts of His‐CBF1 fusion proteins to the IP assays, the amounts of GST‐PIF4 or GST‐PIF5 co‐precipitated with phyB‐GFP were progressively reduced (Fig 5E and F). Together, these data indicated that CBF1 could inhibit phyB interaction with PIF4 and PIF5. Therefore, our data demonstrated that CBF1 promoted PIF4 and PIF5 protein accumulation in the light through both transcriptional and post‐translational regulatory mechanisms.
CBF1 regulation of PIF4 and PIF5 protein abundance is modulated by low temperatures
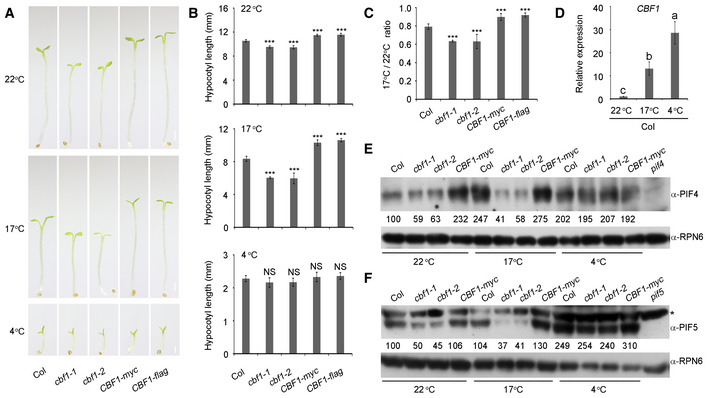
Because CBF1 was shown to play central roles in plant cold acclimation (Jaglo‐Ottosen et al, 1998; Liu et al, 2018; Shi et al, 2018; Ding et al, 2019, 2020), we next asked whether CBF1‐promoted hypocotyl growth was regulated by low temperatures. Wild‐type (Col), two cbf1 mutants, and two CBF1‐OE lines were germinated under long day (LD; 16‐h‐light/8‐h‐dark photoperiod) conditions at 22°C for 2 days, respectively, and then grown under LD conditions at 22°C, or at a lower temperature (17°C), or under cold stress (4°C), for 6 more days. We observed that the hypocotyl growth of all materials was moderately inhibited at 17°C, but severely suppressed at 4°C (Fig 6A and B). After measuring the ratios of hypocotyl lengths at 17°C versus 22°C for the respective genotypes, we found that compared with Col, the two cbf1 mutants displayed increased sensitivity, whereas the two CBF1‐OE lines exhibited decreased sensitivity to hypocotyl growth inhibition at 17°C (Fig 6C). However, Col, cbf1 mutants, and CBF1‐OE lines displayed similar hypocotyl growth inhibition at 4°C (Fig 6A and B). These data suggest that CBF1 attenuates the hypocotyl growth inhibition caused by the lower temperature.
Figure 6. CBF1 regulation of PIF4 and PIF5 protein abundance is modulated by low temperatures.
-
APhenotypes of Col, two cbf1 mutants (cbf1‐1 and cbf1‐2), and two CBF1‐OE lines (CBF1‐myc and CBF1‐flag) grown under LD conditions at 22°C, 17°C and 4°C, respectively. The seeds of different genotypes were germinated under LD (16‐h‐light/8‐h‐dark photoperiod) conditions at 22°C for 2 days, respectively, and then grown under LD conditions at 22°C, 17°C, or 4°C, for additional 6 days. Scale bar = 1 mm.
-
BHypocotyl lengths of Col, two cbf1 mutants (cbf1‐1 and cbf1‐2), and two CBF1‐OE lines (CBF1‐myc and CBF1‐flag) grown under LD conditions at 22°C, 17°C, or 4°C. Error bars represent SD from 10 seedlings. ***P < 0.001 (two‐tailed t‐test) for the indicated genotype compared with Col. NS, not significant.
-
CThe ratios of hypocotyl lengths at 17°C versus 22°C for the indicated seedlings grown under LD conditions. Error bars represent SD from 10 seedlings. ***P < 0.001 (two‐tailed t‐test) for the indicated genotype compared with Col.
-
DqRT–PCR assays showing the CBF1 transcript levels in Col seedlings grown under LD conditions at 22°C, 17°C, and 4°C. Col seeds were germinated under LD conditions at 22°C for 2 days and then grown under LD conditions at 22°C, 17°C, or 4°C for additional 6 days. The samples were collected at ZT8 and then subjected to qRT–PCR assays. Error bars represent SD of three technical replicates. Different letters represent significant differences by one‐way ANOVA with Duncan's post hoc test (P < 0.05).
-
E, FImmunoblots showing the protein levels of PIF4 (E) and PIF5 (F) in Col, cbf1‐1, cbf1‐2, and CBF1‐myc seedlings grown under LD conditions at 22°C, 17°C, and 4°C. The seeds of different genotypes were germinated under LD conditions at 22°C for 2 days and then grown under LD conditions at 22°C, 17°C, or 4°C for additional 6 days. The samples were collected at ZT20 and then subjected to immunoblotting. Anti‐RPN6 was used as a sample loading control. The asterisk in (F) indicates a band that cross‐reacted with the anti‐PIF5 antibody. Numbers below the immunoblots indicate the relative band intensities of PIF4 or PIF5 normalized to those of loading control, respectively. The ratio of the first band was set to 100 for each blot.
Source data are available online for this figure.
We then examined how CBF1‐promoted PIF4 and PIF5 protein accumulation was regulated by low temperatures. Our qRT–PCR data showed that the transcript level of CBF1 was much higher (~ 15‐ to 20‐fold) in Col seedlings at 17°C than at 22°C, and even higher at 4°C (Fig 6D). Interestingly, our immunoblot data indicated that PIF4 proteins accumulated to higher levels in Col seedlings grown at both 17 and 4°C than at 22°C (Fig 6E). However, the levels of PIF4 proteins were comparable at 22°C and at 17°C in the two cbf1 mutant seedlings (Fig 6E), indicating that CBF1 was responsible for promoting PIF4 protein accumulation at 17°C. Moreover, we found that PIF4 proteins accumulated to similar levels in cbf1 mutants and CBF1‐OE lines at 4°C (Fig 6E), suggesting that CBF1 may not be involved in regulating PIF4 protein accumulation under cold stress. Similar patterns were also observed for CBF1 regulation of PIF5 protein abundance at 17 and 4°C (Fig 6F). Together, our data demonstrated that CBF1 promoted PIF4/PIF5 protein accumulation and hypocotyl growth at 22°C and 17°C, but not at 4°C, and that the role of CBF1 in promoting PIF4/PIF5 protein accumulation and hypocotyl growth was more prominent at 17°C than at 22°C.
Discussion
In this study, we showed that CBF1, a well‐characterized pivotal transcription factor regulating plant cold acclimation, promoted hypocotyl elongation under ambient temperatures by positively regulating PIF4 and PIF5 protein accumulation (Fig 7). Our immunoblots showed that the steady‐state levels of endogenous PIF4 and PIF5 proteins were higher in Col seedlings grown in continuous W or R light compared with those in the dark (Fig 4A). Moreover, we demonstrated that CBF1 activated PIF4 and PIF5 expression in the light by directly binding to their promoters (Fig EV3) and that CBF1 enhanced the stability of PIF4 and PIF5 proteins by inhibiting their interaction with phyB (Fig 5). Thus, CBF1 promotes PIF4 and PIF5 protein accumulation in the light through both transcriptional and post‐translational regulatory mechanisms (Fig 7). Notably, higher accumulation of PIF4 in R than in darkness was also observed in a recent study (Park et al, 2018). In addition, our data showed that CBF1 protein was barely detectable in the dark, but CBF1 transcript and protein levels were induced in the light, and interestingly, this induction was mediated by phyA and phyB (Figs 2 and EV1). Therefore, in the light, phytochromes repress hypocotyl growth by inducing phosphorylation and degradation of PIF4 and PIF5 (Nozue et al, 2007; Shen et al, 2007; Lorrain et al, 2008); at the same time, phytochromes induce CBF1 accumulation in the light, which in turn promotes PIF4 and PIF5 protein accumulation (Fig 7). It seems likely that this dual regulation of PIF4 and PIF5 by phytochromes could prevent plants from over‐responding to prolonged light exposure.
Figure 7. A working model depicting that CBF1 promotes hypocotyl elongation under ambient temperatures by positively regulating PIF4 and PIF5 protein accumulation in the light.
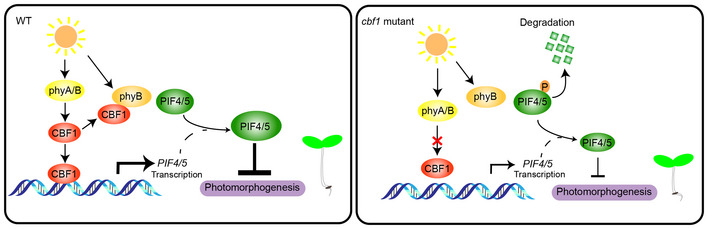
Under ambient temperatures, CBF1 transcript and protein levels are induced by phyA and phyB in the light. CBF1 promotes PIF4 and PIF5 protein accumulation in the light by directly binding to PIF4 and PIF5 promoters to induce their expression, and by inhibiting phyB interaction with PIF4 and PIF5. Thus, CBF1 promotes hypocotyl elongation under ambient temperatures by positively regulating PIF4 and PIF5 protein accumulation in the light through transcriptional and post‐translational mechanisms. Lower levels of PIF4 and PIF5 proteins accumulate in the cbf1 mutants in the light; therefore, cbf1 mutants develop shorter hypocotyls under light conditions.
Interestingly, our data indicated that CBF1 regulation of PIF4/PIF5 protein abundance and hypocotyl growth was modulated by temperatures, with a more prominent role at 17°C than at 22°C (Fig 6). These observations are reminiscent of the fact that Arabidopsis thaliana, both winter and summer annual ecotypes, usually germinate and establish seedlings under relatively low ambient temperatures (spring for summer annuals and fall for winter annuals) (Koornneef et al, 2004). We hypothesize that during the dark‐to‐light transition upon seedlings’ emergence from soil, the role of CBF1 may be to maintain proper hypocotyl growth under low ambient temperatures. This may be essential for seedling establishment and vital for survival of plants in changing natural environments. In support of this hypothesis, studies of natural Arabidopsis populations revealed that in contrast to the various frameshift mutations or nonsynonymous substitutions found in CBF2 and CBF3, almost no frameshift or premature stop codon has been found in CBF1 (Kang et al, 2013; Monroe et al, 2016), suggesting a distinct but pivotal role of CBF1 in plant survival under natural conditions.
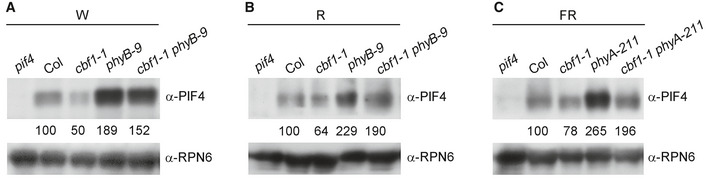
Our genetic data showed that cbf1‐1 phyB‐9 double mutants exhibited intermediate hypocotyl growth phenotypes compared to those of individual single mutants in continuous W and R light (Fig EV2). Similar phenotypes were also observed for cbf1‐1 phyA‐211 in FR light (Fig EV2). The phenotypes of these double mutants might be explained as follows. Firstly, our immunoblots showed that the level of PIF4 in cbf1‐1 phyB‐9 mutants was higher than in Col, but lower than in phyB mutants in continuous W and R light (Fig EV4A and B). PIF4 also accumulated in a similar pattern in Col, phyA‐211, and cbf1‐1 phyA‐211 mutants grown in continuous FR light (Fig EV4C). Thus, the steady‐state levels of endogenous PIF4 proteins might partially explain the hypocotyl lengths of the respective seedlings grown in the light, considering the pivotal role of PIF4 in integrating light and temperature control of hypocotyl growth (Nusinow et al, 2011; Kumar et al, 2012; Sun et al, 2012; Jung et al, 2016; Quint et al, 2016). Secondly, it was previously reported that phyB pif4, phyB pif4 pif5 mutants and phyB pifq mutants also displayed intermediate hypocotyl phenotypes under high R/FR light (de Lucas et al, 2008; Lorrain et al, 2008; Leivar et al, 2012). The phenotypes might be explained by the fact that PIF4 and PIF5 have the intrinsic capacity to induce growth (Nozue et al, 2007; Nusinow et al, 2011; Leivar et al, 2012; Leivar & Monte, 2014). Thus, similar phenotypes of light‐grown cbf1 phyB, phyB pif4, phyB pif4 pif5 and phyB pifq mutants further support our conclusion that CBF1 enhances Arabidopsis hypocotyl growth in the light by promoting PIF4 and PIF5 protein abundance.
Figure EV4. PIF4 protein levels.
-
A–CImmunoblots showing the levels of PIF4 proteins in 4‐day‐old Col, cbf1‐1, phyB‐9, and cbf1‐1 phyB‐9 seedlings grown at 22°C in continuous W (A) or R light (B), and in 4‐day‐old Col, cbf1‐1, phyA‐211, and cbf1‐1 phyA‐211 seedlings grown at 22°C in continuous FR light (C). Numbers below the immunoblots indicate the relative band intensities of PIF4 normalized to those of RPN6, respectively. The ratio was set to 100 for the respective band in Col. The ratio of the first clear band was set to 100 for each blot.
Source data are available online for this figure.
Although it was well documented that the CBF proteins redundantly regulate COR gene expression, distinct roles have also been uncovered for individual CBF proteins. For instance, it was shown that CBF2 negatively regulated the expression of CBF1 and CBF3 in Arabidopsis (Novillo et al, 2004). In addition, comparison of individual cbf single mutants revealed both overlapping and distinct target genes regulated by each CBF protein (Jia et al, 2016; Zhao et al, 2016; Shi et al, 2017), and motif enrichment analysis revealed that more COR genes were direct targets of CBF2 and CBF3 than CBF1 (Shi et al, 2017). Moreover, CBF2 and CBF3 may play more critical roles in the adaptation of Arabidopsis natural populations to different habitats (Alonso‐Blanco et al, 2005; Kang et al, 2013; Oakley et al, 2014; Gehan et al, 2015), while CBF1 may play a more essential role in plant survival under natural conditions based on the fact that almost no frameshift or premature stop codons have been found in CBF1 (Kang et al, 2013; Monroe et al, 2016). In our study, we observed that the cbf1 mutants displayed shorter hypocotyls, whereas the CBF1‐OE lines exhibited longer hypocotyls in all tested light conditions but not in darkness (Fig 1); by contrast, both CBF2 and CBF3‐OE lines showed significant growth retardation in darkness and all light conditions, whereas their single mutants displayed no detectable photomorphogenic phenotypes except that the cbf2 mutants grew longer hypocotyls under blue light (Appendix Fig S2). Our data thus strongly suggest that CBF1 may have evolved distinct roles in regulating plant growth and development compared with CBF2 and CBF3.
It was well documented that the CBF genes were rapidly and dramatically induced by low temperatures (Gilmour et al, 1998; Medina et al, 1999; Vogel et al, 2005; Jiang et al, 2017; Liu et al, 2018; Shi et al, 2018). In this study, our data showed that CBF1 transcript level was also rapidly induced by light (Fig 2C). However, it should be noted that the induction levels of CBF1 by light and cold signals are very different: ~ 10‐ to 20‐fold induction by light (Fig 2C), compared to > 1,000‐fold induction by cold (Jiang et al, 2017). Although the transcription factors involved in cold induction of CBF genes have been extensively studied (Liu et al, 2018; Shi et al, 2018), the molecular mechanisms underlying light induction of CBF1 remain currently obscure. Notably, CBF1 expression rapidly declined after reaching the peak under cold (Jiang et al, 2017) or light treatment (Fig 2C), indicating that CBF1 expression is subjected to strict control by cold and light signals. Interestingly, PIF3, PIF4, and PIF7 were shown to directly bind to the CBF gene promoters and repress their expression under low temperatures (Kidokoro et al, 2009; Lee & Thomashow, 2012; Jiang et al, 2017). Thus, it seems that CBF1 and PIFs form a negative feedback loop where CBF1 activates PIFs but PIFs repress CBF1. It will be fascinating to explore how this regulatory loop is modulated under different temperatures.
Interestingly, the levels of 35S‐driven CBF1 decreased to similar levels in phyA, phyB, and phyA phyB mutants in continuous W and R light (Fig EV1C), suggesting that phyA and phyB play nonredundant and similarly important roles in post‐translational regulation of CBF1 in the light. The fact that CBF1 interacted with only the Pfr form of phyA but with both Pfr and Pr forms of phyB (Fig 3D and E) suggested that CBF1 interacted more selectively with phyA than with phyB in vivo. It will be interesting to investigate whether CBF1 might also regulate the protein stability of PIF1 and PIF3, both of which specifically interacted with the Pfr form of phyA as well (Shimizu‐Sato et al, 2002; Shen et al, 2008). In addition, COP1 expression was shown to be moderately modulated by CBF1 (Appendix Fig S8). Considering the important roles of COP1 in photomorphogenesis (Lau & Deng, 2012), thermomorphogenesis (Delker et al, 2014; Hayes et al, 2017; Park et al, 2017), and cold stress responses (Catalá et al, 2011), the relationship between COP1 and CBF1 under ambient or low temperatures needs to be further characterized. Moreover, given the recent finding that phyB is a thermosensor of ambient temperature (Jung et al, 2016; Legris et al, 2016; Casal & Balasubramanian, 2019), and that PIF4 acts as a central regulator of plant thermomorphogenesis (Koini et al, 2009; Kumar et al, 2012; Quint et al, 2016), it will be intriguing to investigate whether CBF1 is also involved in regulating plant thermomorphogenesis.
Our data showed that CBF1 may not significantly regulate PIF4 and PIF5 protein accumulation at 4°C, although CBF1 expression was strongly induced under this temperature (Fig 6). These observations reflect the complexity of integration mechanisms of light and temperatures. In addition, it should be noted that the steady‐state levels of endogenous PIF4 and PIF5 proteins after prolonged light or temperature treatments were examined in our study, and the patterns may be different from those observed after short light or temperature treatments. For example, PIF4 and PIF5 were shown to be rapidly degraded upon R light exposure (Nozue et al, 2007; Shen et al, 2007; Lorrain et al, 2008), while our data (Fig 4A and C) and those in a previous study (Park et al, 2018) indicated that the steady‐state levels of PIF4 and PIF5 proteins were higher in continuous W or R light compared to those in the dark. Together, our study uncovers an important role of CBF1 in regulating hypocotyl growth under ambient temperatures, thus providing new insights into the tight but complex integration of light and temperature signaling pathways in plants.
Materials and Methods
Plant materials and growth conditions
The wild‐type A. thaliana ecotype used in this study was Columbia (Col), unless otherwise indicated. The phyA‐211 (Reed et al, 1994), phyB‐9 (Reed et al, 1993), phyA‐211 phyB‐9 (Li et al, 2011), pif4‐2 (Leivar et al, 2008a) and pif5‐3 (Khanna et al, 2007), pif4‐101 pif5‐1 (de Lucas et al, 2008), cbf1‐1 (originally named cbf1 in Zhao et al, 2016), cbf1‐2 (originally named cbf1 in Shi et al, 2017), cbf2‐1 (Zhao et al, 2016), cbf2‐2 (Zhao et al, 2016), cbf3‐1 (originally named cbf3 in Shi et al, 2017), cbf3‐2 (originally named cbf3 in Zhao et al, 2016), 35S:PIF4 (Huq & Quail, 2002), and 35S:CBF1‐myc, 35S:CBF2‐myc, and 35S:CBF3‐myc (Liu et al, 2017) were in the Col background and have been described previously. The cbf1 pif4 pif5, 35S:PIF4 cbf1, CBF1‐myc phyA‐211, CBF1‐myc phyB‐9, and CBF1‐myc phyA‐211 phyB‐9 mutants were generated by genetic crosses. The growth conditions and light sources were as described previously (Zhang et al, 2018). The fluence rates of the light growth chambers (Percival Scientific) were 10 μmol m−2 s−1 for continuous white light, 10 μmol m−2 s−1 for continuous far‐red light, 20 μmol m−2 s−1 for continuous red light, and 5 μmol m−2 s−1 for continuous blue light, and 25 μmol m−2 s−1 for white light under long day (16‐h‐light/8‐h‐dark photoperiod) conditions.
Plasmid construction and generation of transgenic Arabidopsis plants
The PHYB‐BD construct was described previously (Zhang et al, 2018). To generate the AD‐PIF4 and AD‐PIF5 constructs, the full‐length coding sequences of PIF4 and PIF5 were amplified by PCR using the respective primer pairs shown in Appendix Table S1 and then cloned into the EcoRI‐XhoI sites of the pGADT7 vector (Clontech). To express CBF1 protein in yeast three‐hybrid assays, the multiple cloning sites of the pRS423 vector (Christianson et al, 1992) were first modified using the primer pair shown in Appendix Table S1 to generate the pRS423‐JL vector. Next, the full‐length coding sequence of CBF1 was amplified by PCR using the primers shown in Appendix Table S1 and cloned into the SacII‐SacI sites of the pRS423‐JL vector.
To generate the constructs expressing GST‐PHYA‐N, GST‐PHYA‐C1, and GST‐PHYA‐C2, the corresponding amplicons were cloned into the BamHI‐SalI sites of the pGEX‐4T‐1 (Amersham Biosciences) vector, respectively. To generate the constructs expressing GST‐PHYB‐N, GST‐PHYB‐C1, and GST‐PHYB‐C2, the corresponding amplicons were cloned into the EcoRI‐NotI (for GST‐PHYB‐N), BamHI‐NotI (for GST‐PHYB‐C1), or BamHI‐XhoI (for GST‐PHYB‐C2) sites of the pGEX‐4T‐1 vector, respectively. To generate the construct expressing GST‐CBF1, the amplicon was cloned into the BamHI‐EcoRI sites of the pGEX‐4T‐1 vector.
To generate the constructs expressing His‐PHYA‐N, His‐PHYA‐C1, and His‐PHYA‐C2, the corresponding amplicons were cloned into the BamHI‐SalI sites of the pET28a (Novagen) vector, respectively. To generate the constructs expressing His‐PHYB‐N, His‐PHYB‐C1, and His‐PHYB‐C2, the corresponding amplicons were cloned into the EcoRI‐NotI (for His‐PHYB‐N), BamHI‐NotI (for His‐PHYB‐C1), or BamHI‐XhoI (for His‐PHYB‐C2) sites of the pET28a vector, respectively. To generate the construct expressing His‐CBF1, the amplicon was cloned into the EcoRI‐XhoI sites of the pET28a vector.
To generate cLuc‐CBF1, the full‐length coding sequence of CBF1 was cloned into the KpnI‐BamHI sites of the 35S:cLuc vector (Chen et al, 2008). To generate phyA‐nLuc, the full‐length coding sequence of PHYA was cloned into the BamHI‐SalI sites of the 35S:nLuc vector (Chen et al, 2008). To generate PIF4‐nLuc and PIF5‐nLuc, the PIF4 and PIF5 amplicons were cloned into the KpnI‐SalI sites of the 35S:nLuc vector (Chen et al, 2008), respectively. To generate cLuc‐PHYB, the PHYB amplicon was cloned into the KpnI site of the 35S:cLuc vector (Chen et al, 2008). To generate the 35S:CBF1‐GFP construct, the CBF1 amplicon was cloned into the HindIII‐SalI sites of the pCAMBIA1300‐GFP vector (Li et al, 2017).
To generate the CBF1p:GUS constructs, the 1.8‐kb promoter fragment of CBF1 was amplified by PCR using the primers shown in Appendix Table S1 and then cloned into the EcoRI and BamHI sites of the pCAMBIA1381 vector.
To generate the 35S:CBF1‐flag construct, the full‐length coding sequence of CBF1 was amplified by PCR using the primers shown in Appendix Table S1 and then cloned into the XbaI and KpnI sites of the pK7FWG2 vector.
To generate various transgenic plants, the corresponding constructs were first transformed into Agrobacterium tumefaciens (strain GV3101) and then into Arabidopsis plants (Col) by the floral dip method (Clough & Bent, 1998).
All of the primers used to generate the constructs mentioned above are listed in Appendix Table S1, and all of the constructs were confirmed by sequencing prior to usage in various assays. Transgenic plants were selected on MS plates in the presence of antibiotics, and homozygous transgenic plants were used in the various assays.
Quantitative RT–PCR (qRT–PCR)
Total RNA was extracted from Arabidopsis seedlings using RNeasy Plant Mini Kits (Tiangen), followed by reverse transcription using RevertAid First Strand cDNA Synthesis Kits (Thermo Fisher Scientific) according to the manufacturers’ instructions. Real‐time PCR was performed using gene‐specific primers and PowerUp SYBR Green PCR Master Mix (Thermo Fisher Scientific). qPCR was performed in triplicate for each sample, and the relative expression levels were normalized to that of a tubulin3 gene. The primers used for qRT–PCR are listed in Appendix Table S1.
GUS staining
CBF1p:GUS transgenic seedlings homozygous for a single copy of the reporter gene were screened as described previously (Wang et al, 2019). Homozygous CBF1p:GUS seedlings were grown in darkness or continuous white, red, far‐red, and blue light for 4 days, and then subjected to GUS staining assays. GUS activity analysis was performed as described previously (Jefferson et al, 1987).
Immunoblotting
For anti‐myc, anti‐phyA, and anti‐phyB immunoblots, total proteins were extracted as described previously (Zhou et al, 2018). Briefly, Arabidopsis seedlings were homogenized in extraction buffer containing 50 mM Tris–HCl, pH 7.5, 10 mM MgCl2, 150 mM NaCl, 1 mM EDTA, 10 mM NaF, 25 mM beta‐glycerophosphate, 2 mM sodium orthovanadate, 10% (w/v) glycerol, 0.1% (w/v) Tween 20, 1 mM PMSF, 1× MG132, and 1× EDTA‐free complete protease inhibitor cocktail (Roche). Protein concentration was measured by the Bradford's assay using Bradford assay reagent (Bio‐Rad), and then, equal amounts of total proteins for each sample were boiled with 6 × SDS loading buffer for 15 min.
For anti‐PIF4 and anti‐PIF5 immunoblots, total proteins were extracted as described previously (Qiu et al, 2017). Briefly, Arabidopsis seedlings were ground in extraction buffer consisting of 100 mM Tris–HCl, pH 7.5, 100 mM NaCl, 5 mM EDTA, pH 8.0, 5% SDS, 20% (w/v) glycerol, 20 mM DTT, 40 mM β‐mercaptoethanol, 2 mM PMSF, 1× EDTA‐free complete protease inhibitor cocktail (Roche), 80 μM MG132 (Sigma), 80 μM MG115 (Sigma), 1% phosphatase inhibitor cocktail (Sigma), and 10 mM N‐ethylmaleimide. Samples were immediately boiled 10 min and then centrifuged for 10 min at 13,000 g at room temperature. Proteins from the supernatants were used in the subsequent immunoblot assays.
Immunoblotting was performed as previously described (Li et al, 2010). Primary antibodies used in this study include anti‐GST (Sigma‐Aldrich), anti‐His (Sigma‐Aldrich), anti‐phyA (Zhang et al, 2018), anti‐myc (Mei5 Biotechnology), anti‐PIF4 (Agrisera), anti‐PIF5 (Agrisera), anti‐RPN6 (Zhou et al, 2018), anti‐HSP (Beijing Protein Innovation), and anti‐H3 (Abcam) antibodies.
The anti‐CBF1 polyclonal and anti‐phyB monoclonal antibodies were made by Beijing Protein Innovation Co, Ltd. (BPI). For generating anti‐CBF1 polyclonal antibodies, 6 × His‐tagged CBF1 fusion proteins were first expressed in E. coli and then purified and used as antigens to immunize rabbits for production of polyclonal antisera. For generating anti‐phyB monoclonal antibodies, His‐phyB‐C2 (900‐1172) proteins expressed in E. coli were used as antigens in mouse to generate the anti‐phyB monoclonal antibodies.
Nuclear protein extraction
Nuclear isolation was performed as described previously (Hetzel et al, 2016). Briefly, 1 g of 4‐day‐old wild‐type (Col) Arabidopsis seedlings grown in darkness or under different light regimes were ground to a fine powder in liquid nitrogen, thawed in 2 ml of pre‐cooled (4°C) grinding buffer (20 mM Tris–HCl, pH 8.0, 35% glycerol, 0.2% Triton X‐100, 5 mM KCl, 5 mM MgCl2, 300 mM sucrose, and 5 mM β‐mercaptoethanol), and filtered twice through a double layer of Miracloth (Merck Millipore). The flow‐through was spun at 5,000 g for 10 min at 4°C, and the pellet, containing the nuclear fraction, was washed four times with 1 ml of grinding buffer and centrifuged at 5,000 g for 10 min at 4°C. The pellet was resuspended in 80 μl of pre‐cooled lysis buffer (10 mM EDTA, pH 8.0, 50 mM Tris–HCl, pH 8.0, and 1% SDS). The final nuclear pellet was resuspended in 6× SDS loading buffer and subjected to immunoblotting. Anti‐H3 was used as a nuclear marker.
Co‐IP
For Co‐IP experiments, homozygous CBF1‐myc transgenic seedlings were first grown in FR or R light for 4 days and then harvested and homogenized in 2 ml of protein extraction buffer and centrifuged twice at 12,000 g for 15 min at 4°C. Then, the extracts were equally divided into two parts and treated with the indicated combinations of R/FR light pulses. Of the 1 ml of supernatant for each sample, 100 μl was reserved as total, and the remainder was incubated with anti‐c‐myc Affinity Gel (Sigma‐Aldrich) for 2 h at 4°C. The beads were then washed three times with protein extraction buffer, and the immunoprecipitated proteins were analyzed by immunoblotting.
In vitro pull‐down assays
For in vitro binding, 2.5 μg of purified recombinant bait proteins (GST‐phyA‐N, GST‐phyA‐C1, GST‐phyA‐C2, GST‐phyB‐N, GST‐phyB‐C1, GST‐phyB‐C2, or GST) and 2.5 μg of prey proteins (6 × His‐CBF1) were added to 500 μl of binding buffer containing 50 mM Tris–HCl, pH 7.5, 100 mM NaCl, 0.2% glycerol, and 0.6% Triton X‐100. After incubation at 4°C for 2 h, Glutathione Sepharose 4B beads (GE Healthcare) were added and incubated for another 2 h. After washing six times with binding buffer, pulled‐down proteins were eluted in 2× SDS loading buffer at 95°C for 15 min, separated on 10% SDS–PAGE gels, and detected by immunoblotting.
LCI assays
Transient LCI assays in tobacco (Nicotiana benthamiana) were performed as described previously (Chen et al, 2008). Briefly, A. tumefaciens (strain GV3101) bacteria containing indicated combinations of constructs were infiltrated into young but fully expanded leaves of 7‐week‐old tobacco plants using a needle‐less syringe. After infiltration, plants were grown under 16‐h‐light/8‐h‐dark for 2 days. Before imaging, the abaxial sides of leaves were sprayed with 1 mM luciferin, and a CCD camera (1300B; Roper) was used to capture the LUC signal at −110°C with 10‐min exposures.
Transcriptome analyses
Total RNA was extracted using the same procedure as for qRT–PCR analysis. Sequencing was performed using the Illumina HiSeq 2000 platform, and the resulting reads were mapped to the reference genome of A. thaliana (TAIR10) with TopHat (Kim et al, 2013; http://tophat.cbcb.umd.edu). Transcript expression was evaluated by cuffdiff (Trapnell et al, 2013; http://cufflinks.cbcb.umd.edu), and transcript abundance was estimated as fragments per kilobase of exon model per million mapped fragments (FPKM). Differentially expressed genes were selected using Student's t‐test with P < 0.05 and fold change > 2.
ChIP
Wild‐type (Col) and CBF1‐myc seedlings grown 4 days under continuous white light were used for ChIP assays following the procedure described previously (Lee et al, 2007). Briefly, 4 g of seedlings were first cross‐linked with 1% formaldehyde under vacuum to cross‐link the protein–DNA complexes. The samples were ground to powder in liquid nitrogen, and the chromatin complexes were isolated and sonicated, and then incubated with anti‐c‐myc Affinity Gel. The precipitated DNA was recovered and analyzed by real‐time qPCR using the respective primer pairs listed in Appendix Table S1. PCRs were performed in triplicate for each sample, and the ChIP values were normalized to their respective DNA input values.
EMSAs
EMSAs were performed using biotin‐labeled probes and the LightShift Chemiluminescent EMSA Kit (Thermo Fisher) according to the manufacturer's instructions with minor modifications. Briefly, 2 μg of purified 6 × His‐CBF1 proteins were incubated together with biotin‐labeled probes in 20 μl reaction buffer. The binding reactions were allowed to proceed at 25°C for 20 min in a thermal cycler (Bio‐Rad) and then separated on 6% native polyacrylamide gels in TBE buffer. The sequences of the complementary oligonucleotides used to generate the biotin‐labeled probes are shown in Appendix Table S1.
Yeast three‐hybrid assays
For yeast three‐hybrid assays, the combinations of constructs to express bait (phyB‐BD), prey (AD‐PIF4 or AD‐PIF5), and CBF1 proteins were co‐transformed into Y190 yeast cells, and the transformants were selected on SD/‐Trp‐Leu‐His agar plates. The yeast cultures were cultivated overnight in SD/‐Trp‐Leu‐His liquid medium supplemented with 2% glucose, and then, 0.5 ml of the overnight cultures were transferred into 1.5 ml liquid SD/‐Trp‐Leu‐His medium supplemented with 25 μM PCB, 2% Galactose, and 1% Raffinose, and cultured with shaking in darkness for 6 h. The yeast cultures were then irradiated with 5‐min R or 5‐min R immediately followed by 5‐min FR light (R + FR) treatment and incubated for 2 h. Then, for each sample, 0.5 ml of yeast cultures were transferred into 0.5 ml of liquid SD/‐Trp‐Leu‐His medium supplemented with 10 μM PCB, 2% Galactose, and 1% Raffinose, then exposed to R or R + FR light treatments again, and incubated for another 2 h. Yeast transformation was conducted as described in the Yeast Protocols Handbook (Clontech), and the liquid assays using ONPG as substrate were performed as described previously (Sheerin et al, 2015; Zhou et al, 2018).
Semi‐in vivo pull‐down assays
The phyB‐GFP seedlings grown in continuous R light for 4 days were used to extract the protein with extraction buffer containing 1 mM EDTA, 150 mM NaCl, 50 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 0.1% NP‐40, 1× MG132, 1 mM PMSF, and 1× EDTA‐free protease inhibitor cocktail. Binding reactions were performed by adding equivalent amounts of phyB‐GFP protein extract, GST‐PIF4 or GST‐PIF5, and GFP‐trap beads (ChromoTek), and different amounts of His‐CBF1 in 1 ml of 1 × PBS buffer containing 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4. The reaction mixtures were incubated at 4°C for 3 h, and then, the beads were washed six times with 1 ml of 1× PBS buffer containing 0.1% NP‐40. The pulled‐down proteins were eluted in 2× SDS loading buffer at 95°C for 15 min, separated on 8% SDS–PAGE gels, and detected by immunoblotting.
Quantification and statistical analysis
Protein quantification and statistical analysis were conducted as described previously (Qi et al, 2020). Protein quantification was performed using ImageJ. Student's t‐tests were performed in Microsoft Excel. ANOVAs were performed with SPSS statistical software. Different letters represent statistical significances determined by one‐way ANOVA (P < 0.05), and levels that are not significantly different are indicated with the same letter.
Author contributions
JL and XD designed research. XD, YY, BJ, YitS, YJ, YihS, JK, HL, DZ, LQ, RH, SZ, YZ, and XW performed research. JC analyzed the RNA‐sequencing data. JL, SY, DK, and HG discussed and interpreted the data. JL, XD, SY, and WT wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Dataset EV1
Source Data for Expanded View and Appendix
Review Process File
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Acknowledgements
We thank Drs Peter Quail and Chuanyou Li for PIF‐related seeds, and Dr. Jian‐Kang Zhu for CBF‐related seeds. This work was supported by grants from the National Natural Science Foundation of China (31970262, 31872658, and 31770321), the National Key Research and Development Program of China (2017YFD0102001), Beijing Outstanding University Discipline Program, and the Recruitment Program of Global Youth Experts of China.
EMBO Journal (2020) 39: e103630
Contributor Information
Dingming Kang, Email: kdm@cau.edu.cn.
Shuhua Yang, Email: yangshuhua@cau.edu.cn.
Jigang Li, Email: jigangli@cau.edu.cn.
Data availability
The RNA‐sequencing data from this publication have been deposited to the Sequence Read Archive database (https://www.ncbi.nlm.nih.gov/sra/) and assigned the accession number PRJNA608253.
References
- Achard P, Gong F, Cheminant S, Alioua M, Hedden P, Genschik P (2008) The cold‐inducible CBF1 factor‐dependent signaling pathway modulates the accumulation of the growth‐repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20: 2117–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso‐Blanco C, Gomez‐Mena C, Llorente F, Koornneef M, Salinas J, Martinez‐Zapater JM (2005) Genetic and molecular analyses of natural variation indicate CBF2 as a candidate gene for underlying a freezing tolerance quantitative trait locus in Arabidopsis . Plant Physiol 139: 1304–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Sady B, Ni W, Kircher S, Schafer E, Quail PH (2006) Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome‐mediated degradation. Mol Cell 23: 439–446 [DOI] [PubMed] [Google Scholar]
- Bae G, Choi G (2008) Decoding of light signals by plant phytochromes and their interacting proteins. Annu Rev Plant Biol 59: 281–311 [DOI] [PubMed] [Google Scholar]
- Bauer D, Viczian A, Kircher S, Nobis T, Nitschke R, Kunkel T, Panigrahi KC, Adam E, Fejes E, Schafer E et al (2004) Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis . Plant Cell 16: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ, Balasubramanian S (2019) Thermomorphogenesis. Annu Rev Plant Biol 70: 321–346 [DOI] [PubMed] [Google Scholar]
- Catalá R, Medina J, Salinas J (2011) Integration of low temperature and light signaling during cold acclimation response in Arabidopsis . Proc Natl Acad Sci USA 108: 16475–16480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM (2008) Firefly luciferase complementation imaging assay for protein‐protein interactions in plants. Plant Physiol 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu J, Zhu JK (2007) Cold stress regulation of gene expression in plants. Trends Plant Sci 12: 444–451 [DOI] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P (1992) Multifunctional yeast high‐copy‐number shuttle vectors. Gene 110: 119–122 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Delker C, Sonntag L, James GV, Janitza P, Ibanez C, Ziermann H, Peterson T, Denk K, Mull S, Ziegler J et al (2014) The DET1‐COP1‐HY5 pathway constitutes a multipurpose signaling module regulating plant photomorphogenesis and thermomorphogenesis. Cell Rep 9: 1983–1989 [DOI] [PubMed] [Google Scholar]
- Ding Y, Lv J, Shi Y, Gao J, Hua J, Song C, Gong Z, Yang S (2019) EGR2 phosphatase regulates OST1 kinase activity and freezing tolerance in Arabidopsis . EMBO J 38: e99819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Shi Y, Yang S (2020) Molecular regulation of plant responses to environmental temperatures. Mol Plant 13: 544–564 [DOI] [PubMed] [Google Scholar]
- Dong CH, Agarwal M, Zhang Y, Xie Q, Zhu JK (2006) The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc Natl Acad Sci USA 103: 8281–8286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Chen M (2008) Transposing phytochrome into the nucleus. Trends Plant Sci 13: 596–601 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Whitelam GC (2007) Light‐quality regulation of freezing tolerance in Arabidopsis thaliana . Nat Genet 39: 1410–1413 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Quail PH (2010) Phytochrome functions in Arabidopsis development. J Exp Bot 61: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD et al (2011) Phytochrome‐interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci USA 108: 20231–20235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii Y, Tanaka H, Konno N, Ogasawara Y, Hamashima N, Tamura S, Hasegawa S, Hayasaki Y, Okajima K, Kodama Y (2017) Phototropin perceives temperature based on the lifetime of its photoactivated state. Proc Natl Acad Sci USA 114: 9206–9211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehan MA, Park S, Gilmour SJ, An C, Lee CM, Thomashow MF (2015) Natural variation in the C‐repeat binding factor cold response pathway correlates with local adaptation of Arabidopsis ecotypes. Plant J 84: 682–693 [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF (1998) Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold‐induced COR gene expression. Plant J 16: 433–442 [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Salazar MP, Everard JD, Thomashow MF (2000) Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol 124: 1854–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Fowler SG, Thomashow MF (2004) Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol Biol 54: 767–781 [DOI] [PubMed] [Google Scholar]
- Hayes S, Sharma A, Fraser DP, Trevisan M, Cragg‐Barber CK, Tavridou E, Fankhauser C, Jenkins GI, Franklin KA (2017) UV‐B perceived by the UVR8 photoreceptor inhibits plant thermomorphogenesis. Curr Biol 27: 120–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzel J, Duttke SH, Benner C, Chory J (2016) Nascent RNA sequencing reveals distinct features in plant transcription. Proc Natl Acad Sci USA 113: 12316–12321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Quail PH (2002) PIF4, a phytochrome‐interacting bHLH factor, function as a negative regulator of phytochrome B signaling in Arabidopsis . EMBO J 21: 2441–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Al‐Sady B, Hudson M, Kim C, Apel K, Quail PH (2004) PHYTOCHROME‐INTERACTING FACTOR 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305: 1937–1941 [DOI] [PubMed] [Google Scholar]
- Jaglo‐Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF (1998) Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280: 104–106 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β‐glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Ding Y, Shi Y, Zhang X, Gong Z, Yang S (2016) The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis . New Phytol 212: 345–353 [DOI] [PubMed] [Google Scholar]
- Jiang B, Shi Y, Zhang X, Xin X, Qi L, Guo H, Li J, Yang S (2017) PIF3 is a negative regulator of the CBF pathway and freezing tolerance in Arabidopsis . Proc Natl Acad Sci USA 114: E6695–E6702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW (2007) Light‐regulated transcriptional networks in higher plants. Nat Rev Genet 8: 217–230 [DOI] [PubMed] [Google Scholar]
- Jung J‐H, Domijan M, Klose C, Biswas S, Ezer D, Gao M, Khattak AK, Box MS, Charoensawan V, Cortijo S et al (2016) Phytochromes function as thermosensors in Arabidopsis . Science 354: 886–889 [DOI] [PubMed] [Google Scholar]
- Kang J, Zhang H, Sun T, Shi Y, Wang J, Zhang B, Wang Z, Zhou Y, Gu H (2013) Natural variation of C‐repeat‐binding factor (CBFs) genes is a major cause of divergence in freezing tolerance among a group of Arabidopsis thaliana populations along the Yangtze River in China. New Phytol 199: 1069–1080 [DOI] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi‐Shinozaki K, Shinozaki K (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress‐inducible transcription factor. Nat Biotechnol 17: 287–291 [DOI] [PubMed] [Google Scholar]
- Khanna R, Huq E, Kikis EA, Al‐Sady B, Lanzatella C, Quail PH (2004) A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix‐loop‐helix transcription factors. Plant Cell 16: 3033–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Shen Y, Marion CM, Tsuchisaka A, Theologis A, Schafer E, Quail PH (2007) The basic helix‐loop‐helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. Plant Cell 19: 3915–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidokoro S, Maruyama K, Nakashima K, Imura Y, Narusaka Y, Shinwari ZK, Osakabe Y, Fujita Y, Mizoi J, Shinozaki K et al (2009) The phytochrome‐interacting factor PIF7 negatively regulates DREB1 expression under circadian control in Arabidopsis . Plant Physiol 151: 2046–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HG, Kim YK, Park JY, Kim J (2002) Light signaling mediated by phytochrome play an important role in cold‐induced gene expression through the C‐repeat/dehydration responsive elements (C/DRE) in Arabidopsis thaliana . Plant J 29: 693–704 [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lee HJ, Jung JH, Lee S, Park CM (2017) HOS1 facilitates the phytochrome B‐mediated inhibition of PIF4 function during hypocotyl growth in Arabidopsis . Mol Plant 10: 274–284 [DOI] [PubMed] [Google Scholar]
- Koini MA, Alvey L, Allen T, Tilley CA, Harberd NP, Whitelam GC, Franklin KA (2009) High temperature‐mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol 19: 408–413 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Rolff E, Spruit CJP (1980) Genetic control of light‐inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Z Pflanzenphysiol 100: 147–160 [Google Scholar]
- Koornneef M, Alonso‐Blanco C, Vreugdenhil D (2004) Naturally occurring genetic variation in Arabidopsis thaliana . Annu Rev Plant Biol 55: 141–172 [DOI] [PubMed] [Google Scholar]
- Kumar SV, Lucyshyn D, Jaeger KE, Alos E, Alvey E, Harberd NP, Wigge PA (2012) Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484: 242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau OS, Deng XW (2012) The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci 17: 584–593 [DOI] [PubMed] [Google Scholar]
- Lazaro A, Mouriz A, Pineiro M, Jarillo JA (2015) Red light‐mediated degradation of CONSTANS by the E3 ubiquitin ligase HOS1 regulates photoperiodic flowering in Arabidopsis . Plant Cell 27: 2437–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Xiong L, Gong Z, Ishitani M, Stevenson B, Zhu JK (2001) The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold‐regulated nucleo‐cytoplasmic partitioning. Genes Dev 15: 912–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C‐M, Thomashow MF (2012) Photoperiodic regulation of the C‐repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana . Proc Natl Acad Sci USA 109: 15054–15059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legris M, Klose C, Burgie ES, Rojas CC, Neme M, Hiltbrunner A, Wigge PA, Schäfer E, Vierstra RD, Casal JJ (2016) Phytochrome B integrates light and temperature signals in Arabidopsis . Science 354: 897–900 [DOI] [PubMed] [Google Scholar]
- Legris M, Ince YC, Fankhauser C (2019) Molecular mechanisms underlying phytochrome‐controlled morphogenesis in plants. Nat Commun 10: 5219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Al‐Sady B, Carle C, Storer A, Alonso JM, Ecker JR, Quail PH (2008a) The Arabidopsis phytochrome‐interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20: 337–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH (2008b) Multiple phytochrome‐interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 18: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Quail PH (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Cohn MM, Quail PH (2012) Phytochrome signaling in green Arabidopsis seedlings: impact assessment of a mutually negative phyB‐PIF feedback loop. Mol Plant 5: 734–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E (2014) PIFs: systems integrators in plant development. Plant Cell 26: 56–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li G, Gao S, Martinez C, He G, Zhou Z, Huang X, Lee JH, Zhang H, Shen Y et al (2010) Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome A signaling. Plant Cell 22: 3634–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li G, Wang H, Deng XW (2011) Phytochrome signaling mechanisms. Arabidopsis Book 9: e0148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ding Y, Shi Y, Zhang X, Zhang S, Gong Z, Yang S (2017) MPK3‐ and MPK6‐mediated ICE1 phosphorylation negatively regulates ICE1 stability and freezing tolerance in Arabidopsis . Dev Cell 43: 630–642 [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi‐Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought‐ and low‐ temperature‐responsive gene expression, respectively, in Arabidopsis . Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Jia Y, Ding Y, Shi Y, Li Z, Guo Y, Gong Z, Yang S (2017) Plasma membrane CRPK1‐mediated phosphorylation of 14‐3‐3 proteins induces their nuclear import to fine‐tune CBF signaling during cold response. Mol Cell 66: 117–128 [DOI] [PubMed] [Google Scholar]
- Liu J, Shi Y, Yang S (2018) Insights into the regulation of C‐repeat binding factors in plant cold signaling. J Integr Plant Biol 60: 780–795 [DOI] [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C (2008) Phytochrome‐mediated inhibition of shade avoidance involves degradation of growth‐promoting bHLH transcription factors. Plant J 53: 312–323 [DOI] [PubMed] [Google Scholar]
- de Lucas M, Daviere JM, Rodriguez‐Falcon M, Pontin M, Iglesias‐Pedraz JM, Lorrain S, Fankhauser C, Blazquez MA, Titarenko E, Prat S (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Medina J, Bargues M, Terol J, Perez‐Alonso M, Salinas J (1999) The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain‐containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol 119: 463–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina J, Catala R, Salinas J (2011) The CBFs: three Arabidopsis transcription factors to cold acclimate. Plant Sci 180: 3–11 [DOI] [PubMed] [Google Scholar]
- Monroe JG, McGovern C, Lasky JR, Grogan K, Beck J, McKay JK (2016) Adaptation to warmer climates by parallel functional evolution of CBF genes in Arabidopsis thaliana . Mol Ecol 25: 3632–3644 [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH (1998) PIF3, a phytochrome‐interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix‐loop‐helix protein. Cell 95: 657–667 [DOI] [PubMed] [Google Scholar]
- Ni W, Xu SL, Chalkley RJ, Pham TN, Guan S, Maltby DA, Burlingame AL, Wang ZY, Quail PH (2013) Multisite light‐induced phosphorylation of the transcription factor PIF3 is necessary for both its rapid degradation and concomitant negative feedback modulation of photoreceptor phyB levels in Arabidopsis . Plant Cell 25: 2679–2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noren L, Kindgren P, Stachula P, Ruhl M, Eriksson ME, Hurry V, Strand A (2016) Circadian and plastid signaling pathways are integrated to ensure correct expression of the CBF and COR genes during photoperiodic growth. Plant Physiol 171: 1392–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novillo F, Alonso JM, Ecker JR, Salinas J (2004) CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis . Proc Natl Acad Sci USA 101: 3985–3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN (2007) Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361 [DOI] [PubMed] [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farre EM, Kay SA (2011) The ELF4‐ELF3‐LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley C, Agren J, Atchison R, Schemske D (2014) QTL mapping of freezing tolerance: links to fitness and adaptive trade‐offs. Mol Ecol 23: 4304–4315 [DOI] [PubMed] [Google Scholar]
- Oh E, Kim J, Park E, Kim JI, Kang C, Choi G (2004) PIL5, a phytochrome‐interacting basic helix‐loop‐helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana . Plant Cell 16: 3045–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund M, Wei N, Deng XW (2000) The roles of photoreceptor systems and the COP1‐targeted destabilization of HY5 in light control of Arabidopsis seedling development. Plant Physiol 124: 1520–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus‐induced development of root and hypocotyl. Genes Dev 11: 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik I, Kathare PK, Kim JI, Huq E (2017) Expanding roles of PIFs in signal integration from multiple processes. Mol Plant 10: 1035–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YJ, Lee HJ, Ha JH, Kim JY, Park CM (2017) COP1 conveys warm temperature information to hypocotyl thermomorphogenesis. New Phytol 215: 269–280 [DOI] [PubMed] [Google Scholar]
- Park E, Kim Y, Choi G (2018) Phytochrome B requires PIF degradation and sequestration to induce light responses across a wide range of light conditions. Plant Cell 30: 1277–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YJ, Lee HJ, Gil KE, Kim JY, Lee JH, Lee H, Cho HT, Vu LD, De Smet I, Park CM (2019) Developmental programming of thermonastic leaf movement. Plant Physiol 180: 1185–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham VN, Kathare PK, Huq E (2018) Phytochromes and phytochrome interacting factors. Plant Physiol 176: 1025–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proveniers MC, van Zanten M (2013) High temperature acclimation through PIF4 signaling. Trends Plant Sci 18: 59–64 [DOI] [PubMed] [Google Scholar]
- Qi L, Liu S, Li C, Fu J, Jing Y, Cheng J, Li H, Zhang D, Wang X, Dong X et al (2020) PHYTOCHROME‐INTERACTING FACTORS interact with the ABA receptors PYL8 and PYL9 to orchestrate ABA signaling in darkness. Mol Plant 13: 414–430 [DOI] [PubMed] [Google Scholar]
- Qiu Y, Pasoreck EK, Reddy AK, Nagatani A, Ma W, Chory J, Chen M (2017) Mechanism of early light signaling by the carboxy‐terminal output module of Arabidopsis phytochrome B. Nat Commun 8: 1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M, Delker C, Franklin KA, Wigge PA, Halliday KJ, van Zanten M (2016) Molecular and genetic control of plant thermomorphogenesis. Nat Plants 2: 15190 [DOI] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J (1993) Mutation in the gene for the red/far red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5: 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagatani A, Elich TD, Fagan M, Chory J (1994) Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol 104: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheerin DJ, Menon C, Oven‐Krockhaus S, Enderle B, Zhu L, Johnen P, Schleifenbaum F, Stierhof YD, Huq E, Hiltbrunner A (2015) Light‐activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27: 189–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Khanna R, Carle CM, Quail PH (2007) Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red‐light activation. Plant Physiol 145: 1043–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Zhu L, Castillon A, Majee M, Downie B, Huq E (2008) Light‐induced phosphorylation and degradation of the negative regulator PHYTOCHROME‐INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20: 1586–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Ding Y, Yang S (2015) Cold signal transduction and its interplay with phytohormones during cold acclimation. Plant Cell Physiol 56: 7–15 [DOI] [PubMed] [Google Scholar]
- Shi Y, Huang J, Sun T, Wang X, Zhu C, Ai Y, Gu H (2017) The precise regulation of different COR genes by individual CBF transcription factors in Arabidopsis thaliana . J Integr Plant Biol 59: 118–133 [DOI] [PubMed] [Google Scholar]
- Shi Y, Ding Y, Yang S (2018) Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci 23: 623–637 [DOI] [PubMed] [Google Scholar]
- Shimizu‐Sato S, Huq E, Tepperman JM, Quail PH (2002) A light‐switchable gene promoter system. Nat Biotech 20: 1041‐1044 [DOI] [PubMed] [Google Scholar]
- Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, Lee CH, Lee D, Choi G (2009) Phytochromes promote seedling light responses by inhibiting four negatively‐acting phytochrome‐interacting factors. Proc Natl Acad Sci USA 106: 7660–7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow AF (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain‐containing transcriptional activator that binds to the C‐repeat/DRE, a cis‐acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA 94: 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Qi L, Li Y, Chu J, Li C (2012) PIF4‐mediated activation of YUCCA8 expression integrates temperature into auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet 8: 1937–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Wang J, Gao Z, Dong J, He H, Terzaghi W, Wei N, Deng XW, Chen H (2016) Arabidopsis SAURs are critical for differential light regulation of the development of various organs. Proc Natl Acad Sci USA 113: 6071–6076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant 50: 571–599 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L (2013) Differential analysis of gene regulation at transcript resolution with RNA‐seq. Nat Biotechnol 31: 46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF (2005) Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis . Plant J 41: 195–211 [DOI] [PubMed] [Google Scholar]
- Vu LD, Xu X, Gevaert K, De Smet I (2019) Developmental plasticity at high temperature. Plant Physiol 181: 399–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Guo C, Peng J, Li C, Wan F, Zhang S, Zhou Y, Yan Y, Qi L, Sun K et al (2019) ABRE‐BINDING FACTORS play a role in the feedback regulation of ABA signaling by mediating rapid ABA induction of ABA co‐receptor genes. New Phytol 221: 341–355 [DOI] [PubMed] [Google Scholar]
- Xu X, Paik I, Zhu L, Huq E (2015) Illuminating progress in phytochrome‐mediated light signaling pathways. Trends Plant Sci 20: 641–650 [DOI] [PubMed] [Google Scholar]
- Yadav A, Singh D, Lingwan M, Yadukrishnan P, Masakapalli SK (2020) Light signaling and UV‐B mediated plant growth regulation. J Integr Plant Biol. 10.1111/jipb.12932 [DOI] [PubMed] [Google Scholar]
- Zhang S, Li C, Zhou Y, Wang X, Li H, Feng Z, Chen H, Qin G, Jin D, Terzaghi W et al (2018) TANDEM ZINC‐FINGER/PLUS3 is a key component of phytochrome A signaling. Plant Cell 30: 835–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Zhang Z, Xie S, Si T, Li Y, Zhu JK (2016) Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis . Plant Physiol 171: 2744–2759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Yang L, Duan J, Cheng J, Shen Y, Wang X, Han R, Li H, Li Z, Wang L et al (2018) Hinge region of Arabidopsis phyA plays an important role in regulating phyA function. Proc Natl Acad Sci USA 115: E11864–E11873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Wu Z, Cao G, Li J, Wei J, Tsuge T, Gu H, Aoyama T, Qu LJ (2014) TRANSLUCENT GREEN, an ERF family transcription factor, controls water balance in Arabidopsis by activating the expression of aquaporin genes. Mol Plant 7: 601–615 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Dataset EV1
Source Data for Expanded View and Appendix
Review Process File
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Data Availability Statement
The RNA‐sequencing data from this publication have been deposited to the Sequence Read Archive database (https://www.ncbi.nlm.nih.gov/sra/) and assigned the accession number PRJNA608253.


