Figure S4.
Theoretical and Experimental Assessment of Several Assumptions Made in the Mathematical Model, Related to Figure 3
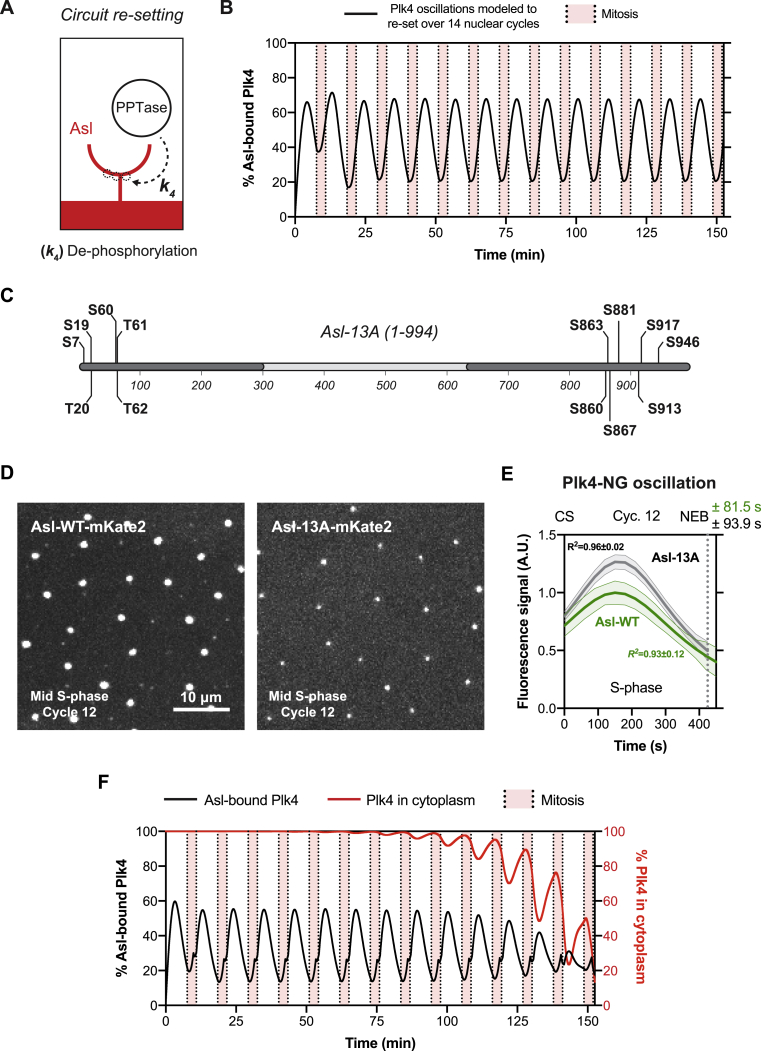
(A) Our mathematical model depicted in Figures 3A and 3B only discretely examines the Plk4-NG oscillation during S-phase of each nuclear cycle. The schematic here shows our speculation that a phosphatase normally removes the phosphate groups (dotted circles) from Asl (red) during mitosis to reset the system for the next oscillation at rate k4 (dotted black arrow).
(B) We implemented this step to extend the original model and plotted the mathematical solution for the percentage of Asl-bound Plk4 molecules (black curve) for a total of 14 nuclear cycles. For simplicity we kept the length of S-phase and mitosis constant through all 14 cycles (see STAR Methods for further details of this extended model).
(C) Schematic shows the Serine (S) and Threonine (T) residues (in bold) that were mutated to Alanine in the Asl-13A construct. Dark gray boxes show the relative positions of the previously mapped Plk4-interacting regions within the N-terminal (Dzhindzhev et al., 2010) and C-terminal (Klebba et al., 2015) regions of Asl.
(D) Micrographs show images from time-lapse movies of embryos expressing Asl-WT-mKate2 and Asl-13A-mKate2 (under the control of their own promoters in an asl mutant background), respectively.
(E) Graphs show the regression data (solid lines) for Plk4-NG oscillations in cycle 12 in embryos expressing either Asl-WT (green) or Asl-13A (dark gray) (both without any fluorescent tag) simultaneously with Plk4-NG. N ≥ 25 embryos for each condition; n = 71 and 68 centrioles (mean) per embryo in Asl-WT or Asl-13A, respectively (collection of two trials performed by two independent researchers, blinded for each other’s data). Data are presented as Mean ± SEM R2 values indicate goodness-of-fit for the regressions. CS = Centrosome separation; NEB = Nuclear envelope breakdown.
(F) In (B) it is assumed that the cytosolic concentration of Plk4 is kept constant over all cycles. The graph here plots an alternative model where the total number of Plk4 molecules in the embryo is kept constant at all cycles. The number of centrioles doubles each cycle, and the mathematical solution for the percentage of Asl-bound Plk4 molecules (black curve), and the percentage of Plk4 molecules that remain in the cytoplasm (red curve), is depicted over 14 nuclear cycles (see STAR Methods for further details and implications of this model). For the first few nuclear cycles, almost all of the Plk4 remains in the cytoplasm since there are only a few centrioles. In the later cycles, however, the amount of Plk4 sequestered by the Asl receptors increases exponentially, as the number of centrioles increase by a factor of 2 in each cycle. Therefore, the rate at which the Asl receptors are able to recruit Plk4 from the cytoplasm decreases, resulting in a reduction in the amplitude of the Plk4 oscillation. This aspect of the model is consistent with our experimental observations that the amplitude of the Plk4 oscillation decreases at later cycles (Figure 1), as does the cytosolic concentration of Plk4 (Figure 3E). An alternative, or additional, mechanism that might explain these observations is that the Plk4 molecules activated by binding to Asl may be more likely to autophosphorylate to stimulate their degradation, so ensuring that more Plk4 is degraded at each cycle as the number of centrioles increase. Interestingly, in either of these scenarios, increasing centriole numbers leads to increasing Plk4 depletion from the cytosol, potentially allowing embryos to effectively “count” their centrioles.