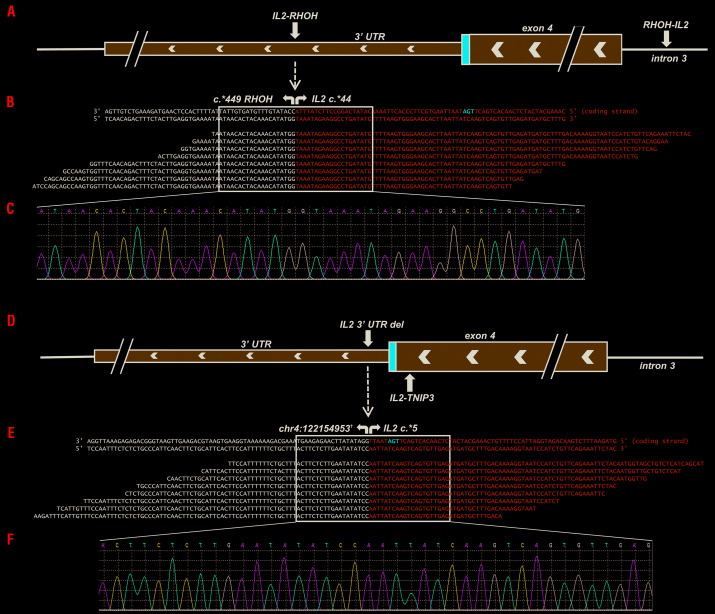
Figure 3.
Structural chromosome alterations of the IL2 gene in CD8+ indolent T-cell lymphoproliferative disorders. In case 7, (A) two chromosome breaks were detected as a consequence of a rearrangement involving the 3′ untranslated region (UTR) of IL2 and 3′ UTR of RHOH (“IL2-RHOH”) and a reciprocal rearrangement involving intron 3 of IL2 and the 3′ UTR of RHOH (“RHOH-IL2”). (B) Pile-up of a subset of reads mapping to the IL2-RHOH rearrangement. (C) Sanger sequencing validation of the fusion breakpoints. In case 8, (D) two chromosome breaks were observed due to a 1.2 Mb deletion spanning the majority of the 3′ UTR of IL2 and a portion of the intergenic region between IL2 and TNIP3 (“IL2 3′ UTR del”) and an inversion involving exon 4 of IL2 and intron 2 of TNIP3 (“IL2-TNIP3”). (E) Pile-up of a subset of reads mapping to the IL2 3′ UTR deletion. (F) Sanger sequencing validation of the deletion breakpoints. †Chromosome position based on assembly GRCh37.p13.