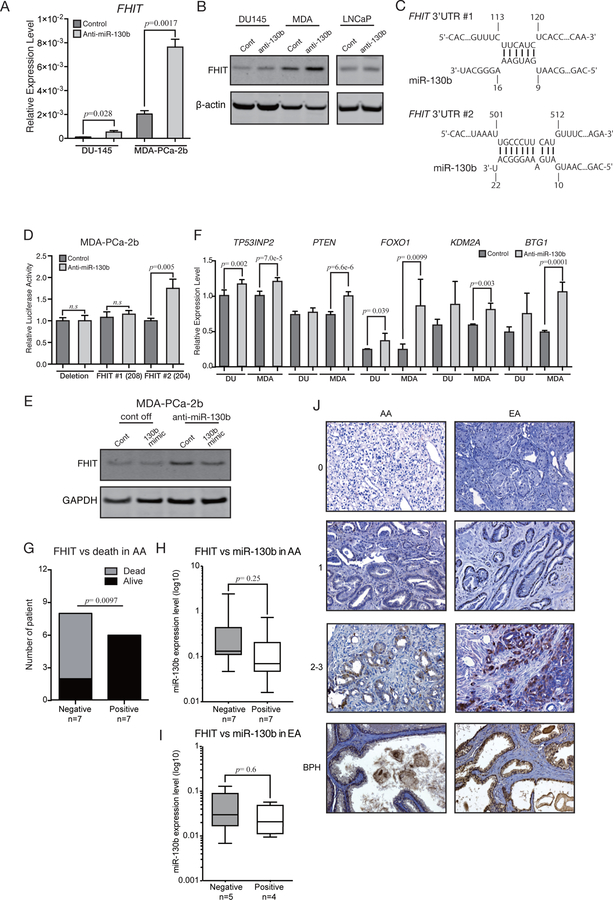
Figure 5. miR-130b affects multiple pathways related to tumor metastasis in AA patients.
(A) qPCR validation for FHIT mRNA levels in stable anti-miR-130b cells (paired t-test; n=3). (B) FHIT Western blotting with stable anti-miR-130b PC cells. (C) FHIT binding sites for miR-130b predicted by IntaRNA. The numbers shown in the panel indicate the position from the 3’UTR start sites. (D) Luciferase reporter assay using vectors containing FHIT binding site or deletion site (paired t-test; n=3). (E) miR-130b rescue experiments to validate that FHIT up-regulation is due to knockdown of miR-130b (cont off, control vector stable MDA-PCa-2b cells; anti-miR-130b, anti-miR-130b stable MDA-PCa-2b cells; cont, cells transfected with negative control #1; 130b mimic, cells transfected with miR-130b mimic). (F) qPCR analysis for reported and predicted miR-130b target genes. RNA samples were isolated from two independent batches of control and anti-miR-130b stable DU-145/ MDA-PCa-2b cells. Then each triplicate well was amplified and measured by qPCR. Each of six expression values were compared by paired t-test (Error bar; mean±SD, n=3). (G) The results of Fisher’s exact test analyzing independency between FHIT expression and SFVAMC AA PC patient death (n=14). (H) The boxplot shows the miR-130b expression level in FHIT negative and positive SFVAMC AA and (I) EA patients. (J) Representative FHIT IHC results in SFVAMC AA, EA patients and BPH tissues. The numbers shown left of the images indicate staining intensity scores.