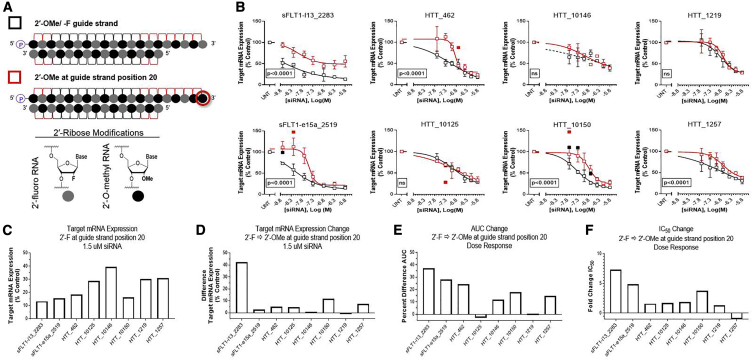
Figure 2.
2′-O-Methyl at 3′ Termini of 20-mer Guide Strands Decreases Activity of Fully Modified, Asymmetric siRNAs
(A) Schematic representations of chemical modification scaffolds used; symbols next to each schematic are used in graphs shown in (B). Legend shows corresponding chemical structures for 2′-ribose modifications. (B) Dose response results (n = 3, mean ± SD). Target name and start site of target sequence are indicated in each graph. HeLa cells were treated with siRNAs at concentrations shown for 72 h. mRNA levels were measured using the Dual-Glo Luciferase Assay System (sFLT1-i13, sFLT1-e15a) or Quantigene 2.0 RNA Assay (HTT) and calculated as a percent of those from untreated cell controls. Statistical outliers were excluded from analyses but are shown in the graphs as solid data points. Nonlinear regression curves with R2 < 0.8 are displayed as dashed rather than solid lines. p values displayed on each graph were calculated by two-way ANOVA. (C) Target mRNA expression with 1.5 μM of each siRNA with 2′-F at guide strand position 20. (D) Differences in target mRNA expression with 1.5 μM of each siRNA when 2′-OMe replaces 2′-F at guide strand position 20. (E) Percent differences in AUCs. (F) Fold changes in IC50 values, calculated by dividing the value obtained from siRNAs with 2′-OMe at guide strand position 20 by the value obtained from siRNAs with 2′-F at guide strand position 20; if this number was <1, then the negative reciprocal is shown. (D–F) Positive or negative values indicate increases or decreases in values when 2′-OMe replaces 2′-F at guide strand position 20.