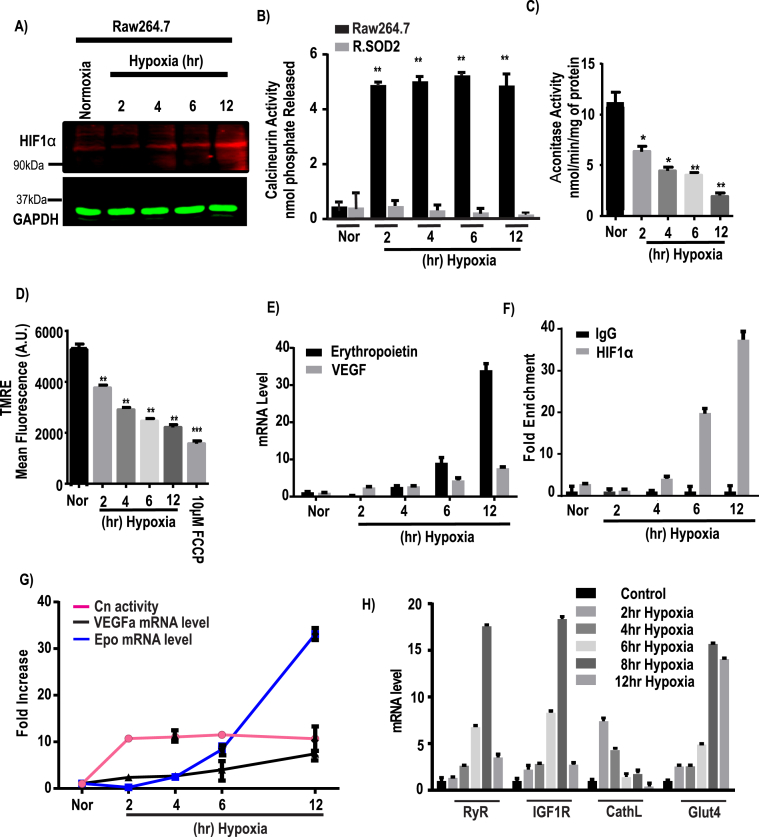
Fig. 1.
Downstream effects of HIF1α and calcineurin A in response to hypoxia. Raw264.7 cells were grown under hypoxia for different time points (0–12h) and each group was divided into two subgroups for immunoblot and biochemical assays. A, a representative immunoblot analysis for HIF1α and GAPDH as loading control, using 40 μg of cell extract in each case. B, calcineurin activity was assayed by using an assay kit as described in the Materials & Methods. C, Aconitase activity assay was performed with 200 μg of total cell lysates as an indicator of oxidative stress and expressed as nmol/min/mg of protein. D, mitochondrial membrane potential ( was measured using TMRE and presented as mean fluorescence intensity (A.U.). E, Hypoxia induced changes in erythropoietin (Epo) and VEGFα mRNA levels. The mRNA levels were monitored by qRT-PCR analysis. Results from 3 independent experiments (n = 3) were normalized to β-actin mRNA levels and expressed as a fold change over the normoxic control. F, Chromatin immunoprecipitation (ChIP) analysis was performed on a HIF1α binding site located on VEGFα promoter at nucleoside −944 to −831 from the transcription start site and enrichment of HIF1α on VEGFα promoter was measured in Raw cells after hypoxic stimulation (n = 3 for all groups). Data represent mean ± S.D. G, the graph represents the fold increase of Cn activity (means ± S.D., n = 3) relative to VEGFα and Epo mRNA levels at each time point of hypoxia after normalization to control. H, mRNA expression profile of different MtRS signaling markers at different time of hypoxia. The mRNA levels were measured by qRT-PCR analysis. Results from 3 independent experiments (n = 3) were normalized to beta actin mRNA levels and expressed as a fold change over the normoxic control. Values showing significant differences from control or individual groups are indicated with an asterisk (*p < 0.05, **p < 0.01).