Figure 13.
Neuronal Phenotypes Contributing to Spatial Separation
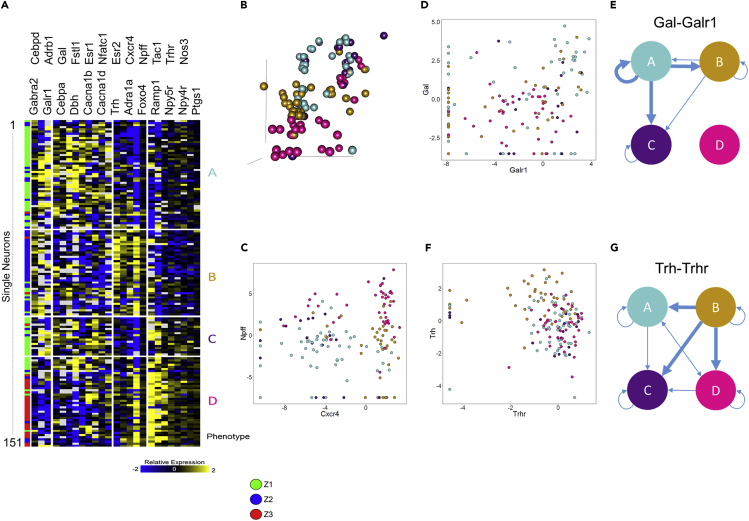
(A) ANOVA was performed with a threshold of 0.001 to find genes that contribute to the separation seen between the three Z groups. Selected genes were clustered using Pearson correlation, resulting in the heatmap shown and giving rise to four different neuronal phenotypes (labeled A–D). We can see that the Z1 group was split into phenotypes A and C, with some samples from the Z1 group clustering along with the Z3 group in phenotype D. Phenotype B is composed mostly of samples residing in the Z2 group.
(B) 3D position of collected samples, colored by phenotype.
(C) Expression of Npff versus Cxcr4, colored for phenotype. We can see that the comparison of these two genes largely determines the phenotypes. Phenotype A is mostly composed of cells that have low expression of Npff and Cxcr4. Phenotype B is composed mostly of cells that have low expression of Npff and high expression of Cxcr4. Phenotype C is composed largely of cells that have high expression of Npff and low expression of Cxcr4. Phenotype D is composed largely of cells that have high expression of both Npff and Cxcr4.
(D) Expression of Galanin versus its receptor Galr1 and (E) network diagram of putative galanin-mediated connectivity between neuronal phenotypes.
(F and G) (F) Expression of thyrotropin-releasing hormone (Trh) and its receptor Trhr and (G) network diagram of putative Trh-mediated connectivity between neuronal phenotypes.