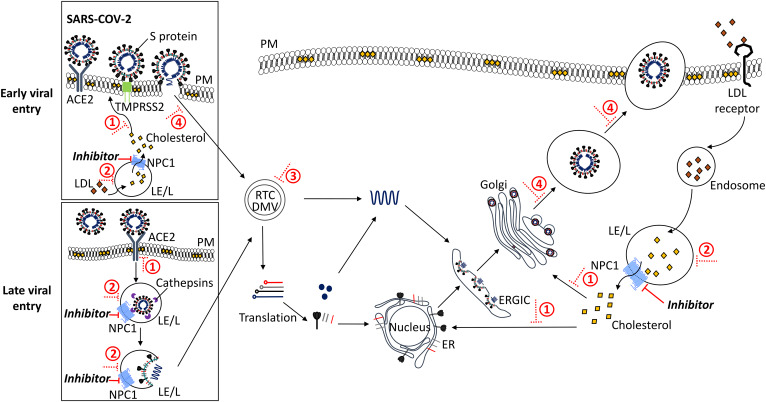
Fig. 1.
Proposed inhibition of SARS-CoV-2 entry/replication by intervention at the NPC1 pathway. Early entry of SARS-CoV-2 into the host cell is mediated by the serine protease, TMPRSS2 (green), which proteolytically primes the viral S protein upon binding to ACE2 (dark gray) at the PM. This releases the virion into the cytoplasm. Alternatively, for late viral entry, SARS-CoV-2 is endocytosed in a clathrin- and/or caveolin-mediated manner from the PM into the host cell. The cysteine protease, cathepsin (purple), cleaves the viral S protein prior to release of the viral nucleocapsid from the LE/L into the cytoplasm. The virus hijacks host-cell machinery to form the replication and transcription complex (RTC) and DMVs where genomic RNA and subgenomic RNA (red, light gray, black, dark blue) are synthesized. Genomic RNA is replicated, and structural and accessory proteins are processed at the ER, before viral assembly at the ER-Golgi intermediate compartment (ERGIC), and bud from the Golgi into vesicles. Finally, the virus is released from the host cell by exocytosis. Under normal conditions, cholesteryl ester in LDL particles (orange) enters the cell by receptor-mediated endocytosis, is hydrolyzed to cholesterol (yellow) in the LE/L where it binds NPC1 (bright blue) and is transported to other organelles (e.g., PM, ER, ERGIC, Golgi). Inhibiting NPC1 (NPC1 inhibitors or the action of NP-C disease mimetics, solid red line) leads to lipid accumulation in the LE/L and, subsequently, 1) depletion of levels of cholesterol in the cell, affecting viral binding and priming, 2) impairment of LE/L pH and protease activity, 3) occurrence of dysregulation at LE/L-ER MCSs, and 4) impairment of endocytosis and exocytosis; overall impeding viral infectivity, replication, assembly, and release (dotted red line).