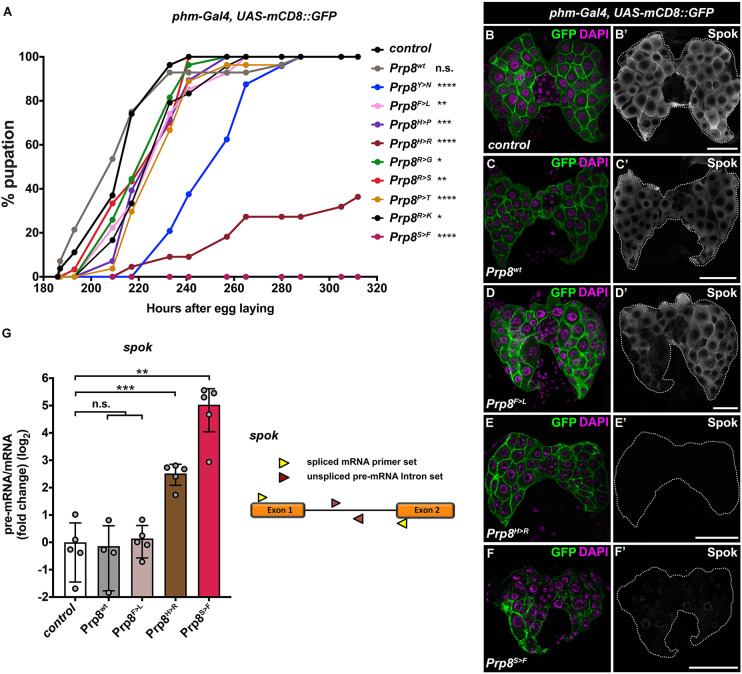
Fig. 3.
Differential impact of RP-Prp8 mutations on the function of Drosophila prothoracic gland. (A) All nine RP-Prp8 mutant variants, but not Prp8wt, delay pupation when overexpressed in the prothoracic gland (PG) using the phm-Gal4 driver. Pupation rates are presented as the percentage of larvae (n≥22 per genotype) that form pupae over time. The pupae were counted at set intervals AEL. Pupation curves represent one of two independent experiments. Statistical significance was determined by Log-rank test. (B–F) Alteration of Spok protein levels in Drosophila PGs (7 days AEL) overexpressing non-tagged RP-Prp8 variants under the control of the phm-Gal4 driver. Relative to control (B′), Prp8wt (C′) and Prp8F>L (D′), the Spok signal was barely detectable in PG glands expressing Prp8H>R (E′) and Prp8S>F (F′). Note the altered morphology of the PG and their nuclei following overexpression of Prp8H>R (E) and Prp8S>F (F). PG cells are highlighted with mCD8::GFP; DAPI stains the nuclei. Panels show projections of multiple confocal sections. Scale bars: 20 µm. (G) PG-specific expression of Prp8H>R and Prp8S>F causes accumulation of unspliced, intron-retaining spok transcript. The pre-mRNA:mRNA ratios shown as a log2 fold-change compared with the control were calculated from the normalized RT-qPCR data by dividing values obtained with intron primer set (red triangles) with values obtained using primers in adjacent exons (yellow triangles). Data are means±s.d., n=4–5. Statistical significance was determined using unpaired t-tests with Welch's correction assuming unequal variance. *P≤0.05, **P≤0.01, ***P<0.001, ****P<0.0001, n.s., non-significant in A and G. The exact number of animals per genotype (A) and biological replicates (G) per sample (n) and P-values are specified in Supplementary Dataset 2.