Fig. 1.
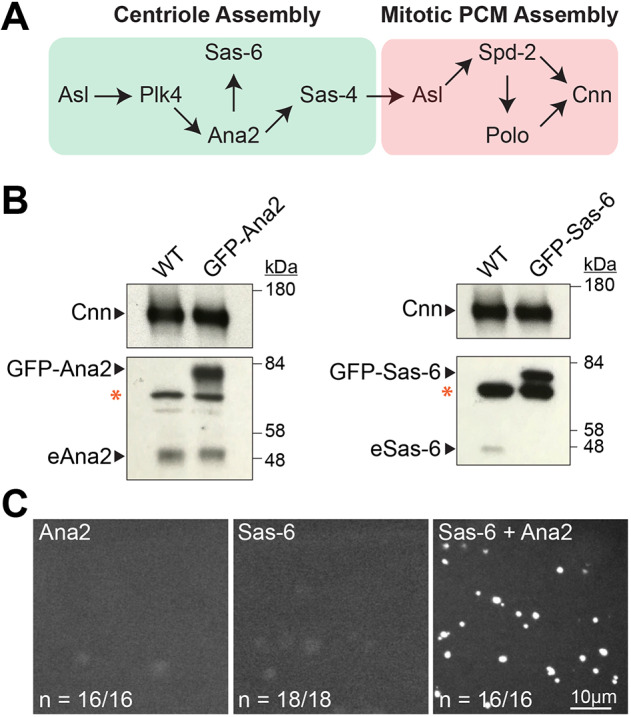
The co-overexpression of Sas-6 and Ana2 leads to SAP formation in Drosophila eggs. (A) Scheme shows putative pathways of centriole assembly (green box) and PCM assembly (pink box) in Drosophila syncytial embryos, illustrating the potential relationship between some of the main proteins involved in these processes. Note that the centrosomes in these rapidly dividing embryos are essentially always in a ‘mitotic’ state (either in mitosis or preparing to enter the next mitosis) and so require Polo, Spd-2 and Cnn to organise this ‘mitotic’ PCM. (B) Western blots of 0- to 3-h-old eggs illustrate the relative expression levels of Ubq-GFP-Ana2 and Ubq-GFP-Sas-6 compared with their endogenous (e) proteins, as indicated; Cnn is shown as a loading control and the red asterisk indicates prominent non-specific bands. Blots are representative examples from two biological repeats. Serial dilution experiments indicate that GFP-Ana2 and GFP-Sas-6 are overexpressed by 3–5× and 5–10× compared with their endogenous proteins, respectively. (C) Confocal images of 0- to 3-h-old eggs expressing either GFP-Ana2, GFP-Sas-6 or both proteins, as indicated. The fraction of eggs exhibiting the phenotype shown is indicated. Note that the dimly fluorescent objects visible in the eggs overexpressing Ana2 or Sas-6 alone are yolk particles.
