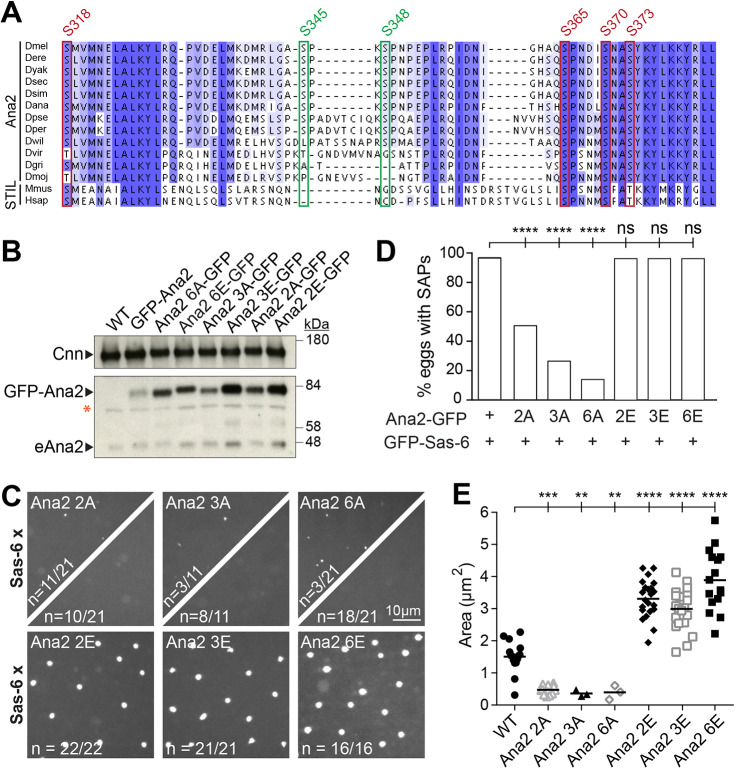
Fig. 6.
Phosphorylation of the Ana2 STAN domain is required for efficient SAP assembly. (A) Multiple sequence alignment of the Ana2/STIL STAN domain in several Drosophila species, mouse and human. Highly conserved Ser/Thr residues are boxed in red, less-well-conserved Ser/Thr residues are boxed in green; numbers above the boxes indicate the position of the indicated Ser residue in the D. melanogaster protein. (B) Western blot illustrates the expression levels in eggs of various WT and mutant Ana2 fusions to GFP compared with each other and with the endogenous Ana2 (eAna2); Cnn is shown as a loading control. The red asterisk indicates a non-specific band. (C) Confocal images of 0- to 3-h-old eggs expressing various Ana2-GFP mutant fusion proteins (as indicated at the top of each image) with WT GFP-Sas-6. For the Ala substitution mutants, some eggs formed a small number of small SAPs (top left of split panel), whereas others formed no detectable SAPs (bottom right of split panel); the fraction of eggs exhibiting each phenotype is indicated. (D) Percentage of eggs laid by females of the indicated genotypes that formed SAPs (n=16–21 eggs per genotype). Fisher's exact test was used to assess statistical significance. (E) SAP size in eggs of the indicated genotype. Each data point represents the average SAP size in an individual egg (N=5–106 SAPs per egg; n=3–22 eggs per genotype). All data were normally distributed according to the D'Agostino and Pearson or Shapiro–Wilk normality test. One-way ANOVA was used to assess statistical significance. ns, not significant; **P<0.01, ***P<0.001, ****P<0.0001.