ABSTRACT
Human disorders of the post-squalene cholesterol biosynthesis pathway frequently result in skeletal abnormalities, yet our understanding of the mechanisms involved is limited. In a forward-genetic approach, we have found that a late-onset skeletal mutant, named kolibernu7, is the result of a cis-acting regulatory mutation leading to loss of methylsterol monooxygenase 1 (msmo1) expression within pre-hypertrophic chondrocytes. Generated msmo1nu81 knockdown mutation resulted in lethality at larval stage. We demonstrated that this is a result of both cholesterol deprivation and sterol intermediate accumulation by creating a mutation eliminating activity of Lanosterol synthase (Lss). Our results indicate that double lssnu60;msmo1nu81 and single lssnu60 mutants survive significantly longer than msmo1nu81 homozygotes. Liver-specific restoration of either Msmo1 or Lss in corresponding mutant backgrounds suppresses larval lethality. Rescued mutants develop dramatic skeletal abnormalities, with a loss of Msmo1 activity resulting in a more-severe patterning defect of a near-complete loss of hypertrophic chondrocytes marked by col10a1a expression. Our analysis suggests that hypertrophic chondrocytes depend on endogenous cholesterol synthesis, and blocking C4 demethylation exacerbates the cholesterol deficiency phenotype. Our findings offer new insight into the genetic control of bone development and provide new zebrafish models for human disorders of the cholesterol biosynthesis pathway.
KEY WORDS: Cholesterol, Chondrodysplasia punctata, Lss, Msmo1, Skeletal dysplasia, Zebrafish
Summary: Characterization of three new zebrafish mutants highlights the role of cholesterol biosynthesis genes in skeletogenesis and establishes zebrafish models for disorders of the post-squalene cholesterol biosynthesis pathway, including chondrodysplasia punctata.
INTRODUCTION
The cholesterol biosynthesis pathway is one of the most complex biochemical pathways and consists of over 30 enzymatic steps (Ačimovič et al., 2016; Sharpe and Brown, 2013). It can be divided into two major sections: the pre-squalene pathway, which is involved in isoprenoid synthesis and contains the rate-limiting enzyme of cholesterol biosynthesis, HMG-CoA reductase, and the post-squalene pathway, which is devoted to sterol synthesis (Ačimovič et al., 2016; Sharpe and Brown, 2013) (Fig. S1). At least ten human disorders result from mutations in post-squalene pathway genes (Herman and Kratz, 2012; Porter and Herman, 2011; Rossi et al., 2015). These disorders are characterized by intellectual disabilities, behavioral problems, heart and genital malformations, eye defects, skin conditions and skeletal deformities (Jira, 2013; Porter and Herman, 2011). Skeletogenesis defects vary from mild to severe, and are seen in endochondral bones, which develop through mineralization of cartilage, and intramembranous bones, which develop directly from condensed mesenchymal cells (Kronenberg, 2003; Ornitz and Marie, 2002).
In a subset of the post-squalene pathway disorders, chondrodysplasia punctata (CDP) is observed (Jurkiewicz et al., 2013). This rare skeletal phenotype is characterized by the observation of dot-like calcium deposits, or punctate, within cartilage on radiographs (Irving et al., 2008; Jurkiewicz et al., 2013; Lykissas, 2013). A result of ectopic calcification, the punctate is most often observed at the end of long bones and within cartilage around joints and the vertebral column (Jurkiewicz et al., 2013). Our understanding of how mutations in the post-squalene cholesterol biosynthesis pathway lead to abnormal skeletogenesis and CDP is limited.
Here, we show that the loss of methylsterol monooxygenase 1 (msmo1) expression within pre-hypertrophic chondrocytes, owing to a cis-acting kolibernu7 (kolnu7) regulatory mutation, results in defective chondrocyte differentiation, irregular bone formation and ectopic ossification within growth plates. We show that loss of Msmo1, and that of Lanosterol synthase (Lss), is lethal in zebrafish larvae. Restoration of hepatic Msmo1 or Lss activity is sufficient for post-larval survival of corresponding mutants. Transgenically rescued mutants develop strong skeletal defects similar to those seen in kolnu7. We show that Msmo1 and Lss activity is needed for proper chondrocyte differentiation, especially in the formation of hypertrophic chondrocytes. Our results suggest that the observed phenotypes are not a result of loss of Indian hedgehog (Ihh) signaling activity within growth plates. The msmo1nu81 mutant phenotype is likely to be the combined result of cholesterol depletion and toxic intermediate accumulation as the lssnu60 mutant, with blocked sterol synthesis, is epistatic to msmo1 and has a less-severe phenotype.
RESULTS
The kolnu7 mutation results in late-onset skeletal deformities
We identified a novel mutation, referred to as kolnu7, based on the reduced body length and small head size of adult homozygote fish (Fig. 1A). The kolnu7 phenotype is first detectable by gross morphological examination at ∼6 weeks of development and the mutation is fully recessive. The kolnu7 homozygote mutants are viable and fertile. Bone and cartilage staining of adult kolnu7 using Alizarin Red and Alcian Blue, respectively (Fig. 1B), reveals defects in both endochondral and intramembranous bones (Bird and Mabee, 2003; Cubbage and Mabee, 1996; Parichy et al., 2009). Specifically, the endochondral bones of adult kolnu7 mutants have dramatically reduced or missing growth plates. Interestingly, only the intramembranous bones located next to cartilaginous elements appear affected. For example, the dentary bone, which develops around Meckel's cartilage (Cubbage and Mabee, 1996), is significantly shortened in mutants relative to wild-type siblings (Fig. S2B,D) and the cartilage itself contains ectopic ossifications (Fig. S2D,D′). Similarly, the observable compressed body phenotype of kolnu7 (Fig. 1A,B) is a result of partial or complete vertebral fusions (Fig. 1B; Fig. S3D,D′). In zebrafish, the vertebrae develop through direct mineralization around a relatively large cartilaginous notochord (Pogoda et al., 2018). In contrast, isolated intramembranous bones, such as the fin rays and operculum, appear relatively unaffected in kolnu7 (Fig. 1B; Fig. S2C). Consistent with the late-onset phenotype of kolnu7, initial cartilage formation and patterning is normal (Fig. S4B), ossification is not prematurely initiated (Fig. 1C-F) and the initial patterning of vertebra centra ossification is normal (Fig. S3A,B).
Fig. 1.
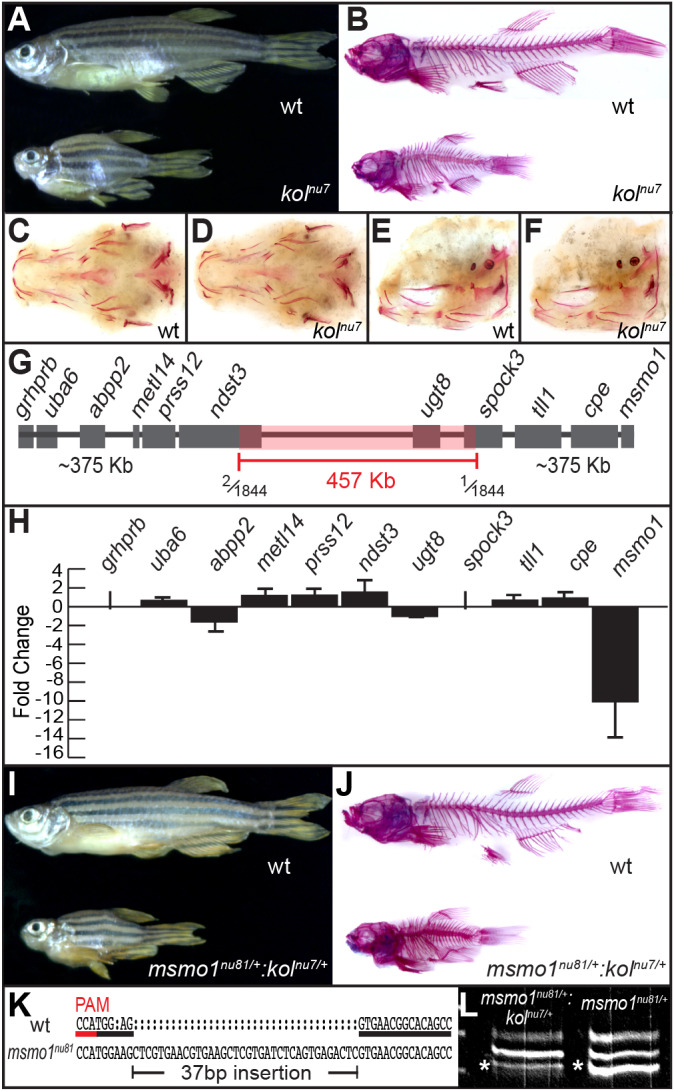
Late-onset skeletal defects observed in the kolibernu7 (kolnu7) mutant are the result of downregulation of msmo1 expression. (A) Compared to wild-type (wt) siblings (top), adult kolnu7 mutants display a reduced body length and small head size. wt n=300, kolnu7 n=300. (B) Whole-mount skeletal preparations reveal gross malformations and hyperossification throughout the adult kolnu7 craniofacial and axial skeleton after Alcian Blue (cartilage) and Alizarin Red (ossified bone and mineralized tissues) staining. wt n=100, kolnu7 n=100. (C-F) Early larval mutants do not display patterning defects or premature ossification. Whole-mount Alizarin Red staining of 4.7 mm (∼8 dpf) wt (C,E) and kolnu7. (D,F). Ventral view (C,D) and lateral view (E,F). wt n=3, kolnu7 n=4. (G) Positional cloning reveals that the kolnu7 locus is located to the ∼457 kb critical region flanked by polymorphic markers with one or two recombinants out of 1844 meioses, corresponding to a genetic distance of 0.16 cM. (H) Screen of gene expression using quantitative RT-PCR from RNA extracted from hypural complex of ∼18 mm SL kolnu7 and wt siblings. wt n=2, kolnu7 =2. Only msmo1 level was significantly different out of 11 tested genes, located in the ∼1.2 Mb region encompassing the kolnu7 locus. Initial screen results: grhprb not detected (ND); uba6 mean difference 0.64, s.d. 0.36; abpp2 −1.62, s.d. 1.00; mettl14 1.16, s.d. 0.68; prss12 1.20, s.d. 0.72; ndst3 1.51, s.d. 1.50; ugt8 −1.03, s.d. 0.07; spock3 ND; tll1 0.67, s.d. 0.27; cpe 0.90, s.d. 0.42; msmo1 −10.07, s.d. 4.32. Confirmation test of msmo1 expression (msmo1 −11.47, s.d. 8.49; P=0.0012), wt n=3, kolnu7 =5. Three technical replicates were included for all assays. Gene expression was normalized to the reference gene eefla1. Fold change was calculated using Livak method (Livak and Schmittgen, 2001). P-value calculated using unpaired Student's t-test on dCt values. (I) The msmo1nu81 mutant allele is not able to complement the kolnu7 mutation. Adult kolnu7/+:msmolnu81/+ transheterozygotes phenocopy the kolnu7 mutant. wt n=100, kolnu7/+:msmolnu81/+ n=100. (J) Whole-mount skeletal preparations reveal gross malformations throughout the kolnu7/+:msmolnu81/+ craniofacial and axial skeleton, similar to those observed in kolnu7. wt n=10, kolnu7/+:msmo1nu81/+ n=10. (K) The msmo1nu81 allele is the result of a 37 bp insertion, allowing for allele-specific expression analysis between msmo1nu81 and kolnu7. PAM, protospacer adjacent motif (underlined in red). (L) Strong downregulation of the kolnu7-linked allele (asterisks) in kolnu7/+:msmo1nu81/+ compared to the wt allele in msmo1nu81/+ suggests that the kolnu7 mutation is cis-acting. kolnu7/+:msmo1nu81/+ n=3, msmo1nu81/+ n=3. The top band is a heterodimer of wt and mutant strands.
Using positional cloning, we mapped the kolnu7 mutation to a critical region located on chromosome 1 flanked by markers segregating in either one or two out of 1844 meioses (Fig. 1G). The critical region physical distance is ∼457 kb, while the genetic distance corresponds to 0.16 cM. As the average physical distance corresponds to ∼650 Mb per 1 cM (Talbot and Schier, 1999), this result indicates a greater than four times reduction in recombination frequency in this region. The critical region overlaps with three known genes (Fig. 1G). Coding sequence analysis of genes within the critical region, as well as those genes neighboring the critical region, did not reveal any changes in protein-coding sequence between wild-type and kolnu7 siblings (data not shown). Using CRISPR/Cas9 genome editing, we mutagenized all three known genes within the critical region and found that each fully complemented the kolnu7 mutation (Table 1; Fig. S5). These results suggested that the kolnu7 mutation disrupts a regulatory sequence located within the critical region.
Table 1.
CRISPR/Cas9 mutations
The kolnu7 mutation negatively regulates expression of msmo1, a gene involved in cholesterol biosynthesis
To test the prediction that the kolnu7 mutation disrupts a cis-acting regulatory element important for bone development, we assessed expression of the genes located in a ∼1.2Mb region containing the kolnu7 locus (Fig. 1G,H). For quantitative real-time PCR (RT-PCR) analysis, we extracted total RNA from the hypural complexes of wild-type and kolnu7 mutant fish. The hypural complex, composed of endochondral bones, is severely affected in kolnu7 mutants. For our analysis, we selected fish that were ∼3 months old, corresponding to a standard length (SL) of ∼18 mm in wild-type siblings. Of the 11 genes analyzed, expression of methylsterol monooxygenase 1 (msmo1) was more than 10-fold downregulated (Fig. 1H). Similarly, using RNA isolated from the cranial vault, which consists of both intramembranous and endochondral bones (Topczewska et al., 2016), we found a 3.8-fold downregulation of msmo1 in kolnu7. Finally, a 2-fold downregulation of msmo1 expression was detected in total RNA isolated from eviscerated trunks of kolnu7 mutants. These results indicate that the kolnu7 mutation induces a downregulation of msmo1 expression, particularly in bone-enriched tissues.
The Msmo1 enzyme catalyzes the removal of a methyl group from C4-methlysterols during the post-squalene cholesterol biosynthesis pathway (Sharpe and Brown, 2013) (Fig. S1). To characterize msmo1 expression in early zebrafish development, we used whole-mount in situ hybridization. Expression was first detected during early stages of somitogenesis in the yolk syncytial layer (YSL) (Fig. 2D), an extraembryonic cell that expresses several markers of the primitive liver (Li et al., 2007; Mudumana et al., 2004). Expression continued in the YSL at 3 days post-fertilization (dpf) (Fig. 2E), with a new domain appearing in the newly formed liver at 4 dpf and 5 dpf (Fig. 2F,F′). Consistent with the late onset of the kolnu7 early phenotype, msmo1 expression was not observed in skeletal elements during the first 5 days of development (Fig. 2E,F). Furthermore, there was no difference in the in situ signals for msmo1 expression between wild-type and kolnu7 siblings during the first 5 days of development (data not shown).
Fig. 2.
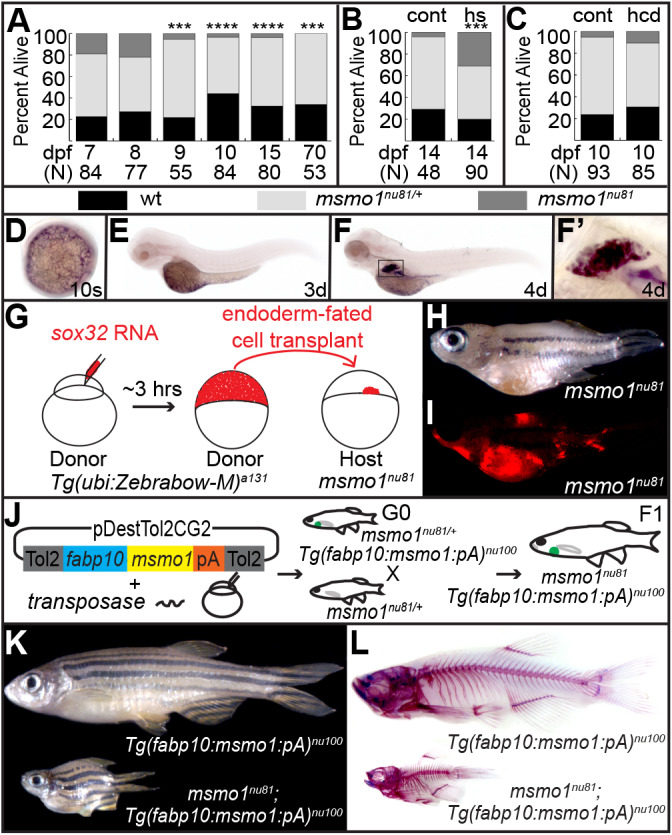
The suppression of early lethality of the msmo1nu81 mutation yields adult fish with kolnu7-like phenotype. (A) Survival of larvae from msmo1nu81/+ in-crosses, from 7 dpf to 70 dpf. Most msmo1nu81 mutants die by 9 dpf. [7 dpf wild type (wt; black) n=19, heterozygotes (het; pale gray) n=49, knockout mutants (KO; dark gray) n=16, P=0.02794; 8 dpf wt n=21, het n=39, KO n=17, P=0.8065; 9 dpf wt n=12, het n=40, KO n=3, ***P=0.0008; 10 dpf wt n=37, het n=44, KO n=3, ****P<0.0001; 15 dpf wt n=26, het n=51, KO n=3, ****P<0.0001; 70 dpf wt n=18, het n=35, KO n=0, ***P=0.001.] (B) Overexpression of msmo1 driven by daily heat shock of the transgenic line Tg(hsp70l:msmo1:IRESnlsGFP)nu99 rescued the lethality of msmo1nu8 mutants. Transgenic screening based on cardiac GFP. (Control non-transgenic siblings 14 dpf wt n=14, het n=32, KO n=2, ***P=0.0035; transgenic siblings 14 dpf wt n=18, het n=44, KO n=28, P=0.3214.) cont, control; hs, heat shock. (C) Dietary cholesterol supplementation does not improve the survivability of msmo1nu81 mutants. Clutches from msmo1nu81/+ in-crosses were fed either a high-cholesterol diet (hcd) or a control standardized diet (cont) beginning at 5 dpf until collection at 10 dpf. (HCD wt n=26, het n=50, KO n=9, P=0.0089; control diet wt n=22, het n=66, KO n=5, P<0.0001.) All two-tailed P-values were calculated using chi-squared test. (D-F′) Whole-mount in situ hybridization during the first 5 days of development shows msmo1 expression predominately in the yolk syncytial layer (YSL) and liver. Expression is first detected during early somitogenesis in the YSL (D) and continues there at 3 dpf (E). At 4 dpf, strong expression is observed in the differentiated liver (F,F′). (G-I) Generation of msmo1nu81/wild-type chimeras using endoderm replacement rescues early lethality of msmo1nu81 mutants and reveals a strong kolnu7-like phenotype. (G) Schematic of the procedure. Wild-type Tg(ubi:Zebrabow-M)a131 donor embryos were injected with sox32 RNA to force an endodermal fate. At high stage (∼3 hpf), cells were transplanted from donor to host embryos collected from msmo1nu81/+ in-crosses. (H) Surviving msmo1nu81 mutants display a strong kolnu7-like phenotype. msmo1nu81 n=4. (I) The majority of organs of endodermal origin displayed a high enrichment in transplanted Tg(ubi:Zebrabow-M)a131 cells (red). msmo1nu81 n=4. (J-L) Liver-specific msmo1 expression in msmo1nu81 mutants rescues early lethality and produces juvenile msmo1nu81 mutants with strong kolnu7-like phenotype. (J) Liver-specific regulatory element fabp10a was used to drive msmo1 expression in msmo1nu81 mutants. (K) Adult Tg(fabp10a:msmo1:pA)nu100;msmo1nu81 mutants phenocopy kolnu7 mutant. Tg(fabp10:msmo1:pA)nu100 n=50, Tg(fabp10a:msmo1:pA)nu100;msmo1nu81 n=50. (L) Whole-mount skeletal preparations reveal gross malformations throughout Tg(fabp10a:msmo1:pA)nu100;msmo1nu81 craniofacial and axial skeleton, similar to those observed in kolnu7. Tg(fabp10:msmo1:pA)nu100 n=3, Tg(fabp10a:msmo1:pA)nu100;msmo1nu81 n=7.
We next characterized msmo1 expression during juvenile development by RNAscope in situ hybridization. In ∼2-month-old fish, SL ∼15 mm, msmo1 expression was found within cartilaginous elements such as Meckel's cartilage, but not within intramembranous skeletal elements such as the dentary bone (Fig. S6H) (Bird and Mabee, 2003; Cubbage and Mabee, 1996). Importantly, expression of msmo1 was seen within endochondral growth plates, specifically in the region corresponding to pre-hypertrophic chondrocytes (Kronenberg, 2003; LeClair et al., 2009) (see Fig. 5C,C′). In addition, we observed msmo1 expression in the liver, kidney, intestine, brain, retina, spinal cord and skin (Fig. S6A-G). Comparison of msmo1 expression in juvenile wild-type and kolnu7 siblings revealed an undetectable level of msmo1 expression within endochondral growth plates of kolnu7 mutants (Fig. 5F,F′). Our findings support the notion that a deficit in msmo1 expression, particularly in differentiating chondrocytes, underlies the kolnu7 phenotype.
The kolnu7 mutation perturbs a cis-acting msmo1 regulatory element
To confirm that loss of Msmo1 is solely responsible for the development of the kolnu7 phenotype, we mutagenized the msmo1 gene using CRISPR/Cas9 genome editing. We isolated an allele, msmo1nu81, which results in a 37 bp insertion (Fig. 1K) in the first coding exon, resulting in a frameshift and premature protein truncation. The msmo1nu81 heterozygotes, similar to kolnu7/+, are phenotypically normal. Next, to test genetic complementation, we crossed msmo1nu81 and kolnu7 heterozygotes. Transheterozygote kolnu7/+:msmo1nu81/+ appeared phenotypically normal until ∼5 weeks post-fertilization, after which they began to take on a kolnu7-like appearance and became morphologically indistinguishable from kolnu7 mutants (Fig. 1I,J).
Our results suggest that the kolnu7 mutation disrupts a regulatory element driving msmo1 expression. We directly examined this possibility by taking advantage of a relatively large insertion in msmo1nu81 (Fig. 1K) that allows us to compare allele-specific expression. Once again, we used total RNA isolated from the hypural complexes for semi-quantitative RT-PCR analysis (Fig. 1L). In contrast to msmo1nu81/+, in which both wild-type and mutant products can easily be identified after RT-PCR, the wild-type msmo1 allele linked to the kolnu7 locus is under-represented, strongly supporting the notion that the kolnu7 mutation disrupts a cis-acting regulatory element. In summary, based on the results of our positional cloning, in situ hybridization and complementation testing, we concluded that the kolnu7 mutant phenotype is a result of strongly reduced msmo1 expression after loss of a positively acting regulatory element.
Loss of Msmo1 function is lethal in zebrafish larvae
While kolnu7 homozygote fish survive until adulthood, msmo1nu81 homozygote mutants show a decrease in growth at 6 dpf (data not shown) and die by 9 dpf (Fig. 2A). Overexpression of wild-type msmo1 driven by the Tg(hsp70l:msmo1:IRESnlsEGFP)nu99 transgene was able to suppress the early lethality of msmo1nu81 homozygotes (Fig. 2B), indicating that loss of Msmo1 is responsible for the death of the msmo1nu81 mutants. Because Msmo1 activity plays a crucial role in cholesterol biosynthesis (Sharpe and Brown, 2013) (Fig. S1), we tested whether a cholesterol-enriched diet could extend the lifespan of msmo1nu81 mutants. However, even a 4% cholesterol-enriched diet, shown to induce hypercholesterolemia in zebrafish (Stoletov et al., 2009), did not significantly improve mutants’ survival (Fig. 2C).
Expression of msmo1 in liver rescues msmo1nu81 mutants and produces kolnu7 phenotype
To study the role of Msmo1 in bone formation, we needed to suppress the early lethality of msmo1nu81 mutants. We observed strong msmo1 expression in the larval and juvenile liver (Fig. 2D-F′; Fig. S6A), an organ responsible for producing endogenous cholesterol (Turley, 2004). We therefore predicted that hepatic restoration of Msmo1 activity might rescue the msmo1nu81 mutants, allowing us to study juvenile bone development in this genetic background. To test this prediction, we used two approaches. First, with the help of partial endoderm replacement, we created msmo1nu81 mutants with chimeric endodermal organs. We pushed cells of the Tg(ubi:Zebrabow-M)a131 (Pan et al., 2013) donor embryos to an endodermal fate by overexpressing sox32 at the one-cell stage (Stafford et al., 2006). The red fluorescing donor cells were transplanted from high-stage donor embryos into shield-stage host embryos obtained from msmo1nu81 heterozygote in-crosses (Fig. 2G). We analyzed the host fish at ∼5 weeks of development. Four of 15 transplanted fish, with significant contribution of donor cells, appeared to have a kolnu7-like appearance (Fig. 2H,I). Genotyping revealed that all of the kolnu7-like fish were chimeras with msmo1nu81 mutants. In a parallel effort, we employed a transgenic approach to drive hepatic msmo1 expression using the liver-specific regulatory element of the fatty acid binding protein, liver basic (fabp10a) gene (Her et al., 2003; Kwan et al., 2007) (Fig. 2J). In contrast to the transplantation experiments, the transgenic expression of msmo1 was much more efficient in suppression of lethality, and close to Mendelian ratios of msmo1nu81 mutants were found at 5 weeks in ∼700 fish studied from 13 separate crosses. Rescued mutants grew to adulthood and displayed a severe kolnu7-like phenotype (Fig. 2K,L). These results indicate that restoration of hepatic Msmo1 activity is sufficient for survival of msmo1nu81 mutants beyond early larval stages. Interestingly, liver-derived cholesterol synthesis is unable to rescue the skeletal phenotype of msmo1nu81 mutants, indicating a requirement for chondrocyte-specific sterol production.
Loss of sterol synthesis partially suppresses msmo1nu81 mutant phenotype
In most human syndromes resulting from mutations in genes involved in the post-squalene cholesterol biosynthesis pathway, a simultaneous decrease in cholesterol levels and an accumulation of sterol intermediates is observed (Porter and Herman, 2011). To elucidate the effects of cholesterol deprivation from sterol intermediates’ accumulation, we eliminated the activity of Lss. This enzyme catalyzes the cyclization of the first sterol in the cholesterol biosynthesis pathway (Huff and Telford, 2005) (Fig. S1). Mutation of the lss gene is predicted to result in a deficit of cholesterol biosynthesis, but not an accumulation of sterol intermediates (Sharpe and Brown, 2013). Using CRISPR/Cas9 genome editing, we generated the lssnu60 allele, resulting in a 23 bp deletion (Fig. 3A), causing a frameshift and premature protein truncation. Heterozygote lssnu60 fish are phenotypically normal and fertile. In contrast to kolnu7/+:msmo1nu81/+ transheterozygotes, the study of over 180 fish from two separate crosses did not identify any signs of skeletal abnormalities in adult kolnu7/+;lssnu60/+ animals (data not shown). This observation further supports the notion that a reduced level of Msmo1 activity, and not a general deficit in the cholesterol biosynthesis pathway, is responsible for the kolnu7 phenotype.
Fig. 3.
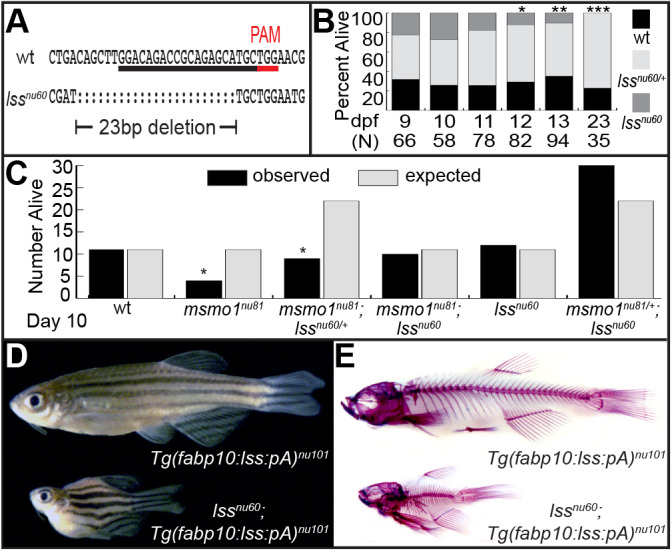
Loss of Lss activity phenotype is epistatic to msmo1nu81 mutation. (A) The lssnu60 allele. PAM, protospacer adjacent motif (underlined in red). (B) Survival analysis of clutches of lssnu60/+ in-crosses, from 9 dpf to 23 dpf, reveals that most lssnu60 mutants die by 12 dpf. [9 dpf wild type (wt; black) n=21, heterozygotes (het; pale gray) n=30, knockout mutants (KO; dark gray) n=15, P=0.4412; 10 dpf wt n=15, het n=27, KO n=16, P=0.8563; 11 dpf wt n=20, het n=44, KO n=14, P=0.3320; 12 dpf wt n=24, het n=48, KO n=10, *P=0.0277; 13 dpf wt n=33, het n=51, KO n=10, **P=0.0026; 23 dpf wt n=8, het n=27, KO n=0, ***P=0.0009.] (C) Survival analysis of clutches of msmo1nu81/+;lssnu60/+ in-crosses reveals that loss of Lss activity increases survivability of msmo1nu81 mutants at 10 dpf. [wt observed (O) n=11, expected (E) n=11; msmo1nu81 O n=4, E n=11; msmo1nu81;lssnu60/+ O n=9, E n=22; msmo1nu81;lssnu60 O n=10, E n=11; lssnu60 O n=12, E n=11; msmo1nu81/+;lssnu60 O n=30, E n=22; msmo1nu81/+;lssnu60/+ O n=40, E n=44; msmo1nu81/+ O n=18, E n=22; lssnu60/+ O n=18, E n=22; *P=0.0431.] Two-tailed P-values were calculated using chi-squared test. (D,E) Suppression of early lethality of lssnu60 mutants by liver-specific expression of lss. (D) Adult Tg(fabp10:lss:pA)nu101;lssnu60 mutants phenocopy kolnu7. Tg(fabp10:lss:pA)nu101 n=12, Tg(fabp10:lss:pA)nu101;lssnu60 n=12. (E) Whole-mount skeletal preparations reveal gross malformations throughout Tg(fabp10:lss:pA)nu101;lssnu60 craniofacial and axial skeleton, similar to those observed in kolnu7. Tg(fabp10:lss:pA)nu101 n=3, Tg(fabp10:lss:pA)nu101;lssnu60 n=5.
The growth of msmo1nu81 and lssnu60 homozygote mutants lags behind that of wild-type siblings beginning at 6 dpf (data not shown). Unlike msmo1nu81 mutants, lssnu60 homozygotes live until 11 dpf (Fig. 3B). A measurement of total cholesterol levels at 8 dpf indicated a strong reduction in both mutants. Compared to wild-type siblings, total cholesterol levels of lssnu60 homozygous mutants were 43% (s.d. 9.14, P=0.0011) of wild-type levels, whereas those of msmo1nu81 homozygous mutants were 22% (s.d. 10.59, P=0.0006) of wild-type levels. To determine whether better survival is a result of a less-severe defect in cholesterol biosynthesis or an accumulation of sterol intermediates, we conducted an epistatic analysis. We found that double lssnu60;msmo1nu81 mutants survived to 10 dpf in close to expected numbers, whereas most msmo1nu81 single mutants were lost (Fig. 3C). Our results strongly suggest that an accumulation of sterol intermediates is responsible for the more-severe msmo1nu81 phenotype, compared to that of lssnu60 mutants.
Loss of Msmo1 and Lss activity is associated with skeletal defects
To study juvenile bone development in the lssnu60 knockout mutants, we again employed a transgenic approach to restore hepatic lss expression (Her et al., 2003; Kwan et al., 2007). Similar to the results described for the rescued msmo1nu81 mutants, close to Mendelian ratios of rescued lssnu60 were found at 5 weeks in ∼425 fish studied from nine separate crosses. Rescued mutants grew to adulthood and displayed a severe kolnu7-like phenotype (Fig. 3D). Skeletal staining revealed compressions of the axial skeleton due to vertebral column fusions and deformations in the craniofacial skeleton (Fig. 3E).
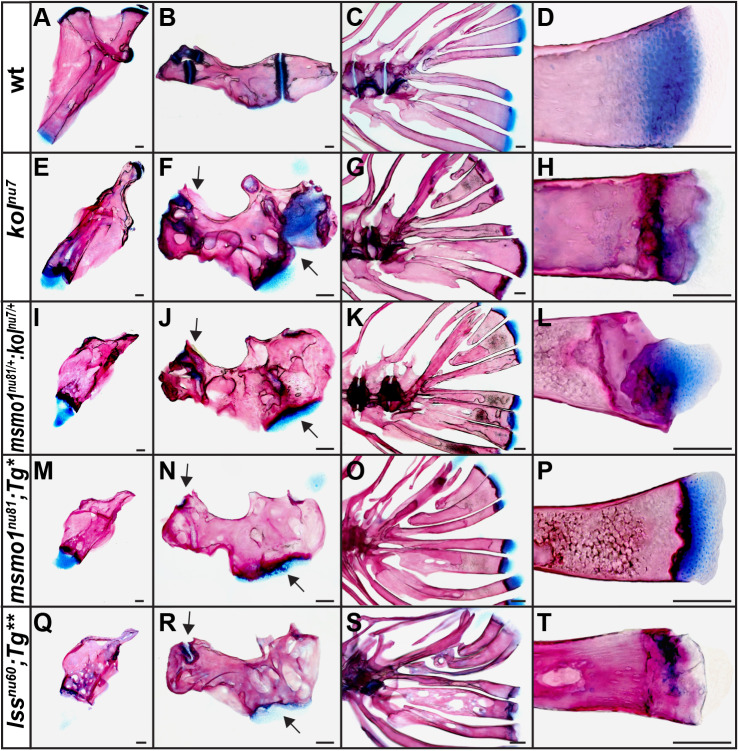
To better understand the skeletal phenotype associated with loss of Msmo1 and Lss activity, we analyzed the skeletons of kolnu7, kolnu7/+:msmo1nu81/+, Tg(fabp10a:msmo1:pA)nu100;msmo1nu81 and Tg(fabp10:lss:pA)nu101;lssnu60 mutants. We collected mutant and wild-type siblings at 4 months and compared bone formation, focusing on selected bones formed by cartilage replacement mechanisms: the hyomandibular, ceratohyal and hypural complex (Bird and Mabee, 2003; Cubbage and Mabee, 1996) (Fig. 4). The bones of mutants and transheterozygotes were irregularly shaped (Fig. 4A,E,I,M,Q) with abnormal stratification of the growth plates (Fig. 4D,H,L,P,T). The persistent cartilage bands separating individual bones (Fig. 4B), which normally never fuse in zebrafish (LeClair et al., 2009), were often closed as a result of ectopic ossification (Fig. 4F,J,N,R). The endochondral bones of the kolnu7/+:msmo1nu81/+ (Fig. 4I-L), Tg(fabp10a:msmo1:pA)nu100;msmo1nu81 (Fig. 4M-P) and Tg(fabp10:lss:pA)nu101;lssnu60 mutants (Fig. 4Q-T) phenocopied those of the kolnu7 mutant (Fig. 4E-H), indicating that the loss of Msmo1 and Lss activity within cartilage-derived elements produces severely deformed, compressed endochondral bones with frequent ectopic ossification of the growth plate.
Fig. 4.
Loss of Msmo1 and Lss activity is associated with abnormal endochondral bone formation. (A-T) The hyomandibular (A,E,I,M,Q), ceratohyal (B,F,J,N,R) and hypural complex (C,D,G,H,K,L,O,P,S,T) were compared between wild type (A-D), kolnu7 (E-H), kolnu7/+:msmolnu81/+ (I-L), Tg(fabp10a:msmo1:pA)nu100;msmo1nu81 (M-P) and Tg(fabp10:lss:pA)nu101;lssnu60 (Q-T). (E-T) The endochondral bones of the mutants are compressed and irregularly shaped with ectopic ossification and fusions throughout the growth plates, marked with arrows. Images represent disarticulated whole-mount adult skeletal preparations at ∼4 months of age. Scale bars: 100μm. Tg*, Tg(fabp10:msmo1:pA)nu100; Tg**, Tg(fabp10:lss:pA)nu101. wt n=4, kolnu7 n=4, kolnu7/+:msmolnu81/+ n=4, Tg(fabp10a:msmo1:pA)nu100;msmo1nu81 n=4, Tg(fabp10:lss:pA)nu101;lssnu60 n=4.
Msmo1 and Lss activity in the growth plate is needed for normal chondrocyte differentiation
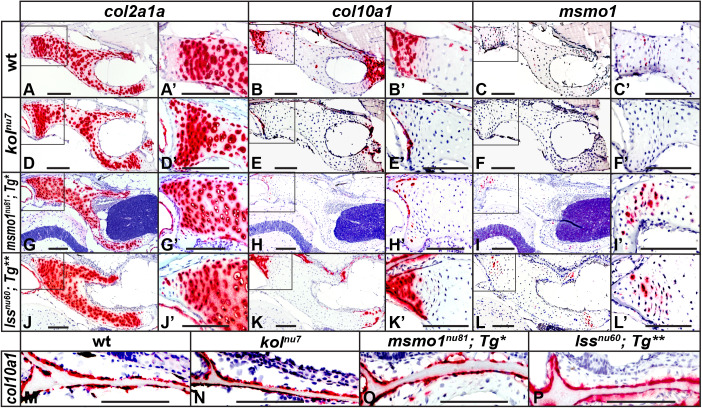
To examine changes in growth plate patterning after loss of Msmo1 and Lss activity, we used RNAscope in situ hybridization to analyze the gene expression of key markers in the growth plate of the pterotic bone, an element of the chondocranium (Cubbage and Mabee, 1996). We focused on collagen type II, alpha 1 (col2a1a), a marker of resting and proliferating chondrocytes (Dale and Topczewski, 2011; Zhao et al., 1997), and collagen type 10, alpha 1a (col10a1a), a marker of hypertrophic chondrocytes (Dale and Topczewski, 2011; Eames et al., 2012; Topczewska et al., 2016) and osteoblasts (Avaron et al., 2006; Eames et al., 2012; Kim et al., 2013). We analyzed wild-type, kolnu7, Tg(fabp10a:msmo1:pA)nu100;msmo1nu81 and Tg(fabp10:lss:pA)nu101;lssnu60 fish at ∼2 months of age, when wild-type fish had an SL of ∼15 mm. In wild-type animals, strong col2a1a expression was observed in round and stacked chondrocytes (Fig. 5A,A′), corresponding to presumed resting and proliferative zones of the growth plate. In a complementary pattern, col10a1a was detected in the presumed hypertrophic chondrocytes (Fig. 5B,B′). In the kolnu7 and Tg(fabp10a:msmo1:pA)nu100;msmo1nu81 mutants, col2a1a expression is expanded (Fig. 5D,D′,G,G′) and accompanied by near complete loss of col10a1a expression (Fig. 5E,E′,H,H′). Although a similar expansion of the of col2a1a domain was observed in Tg(fabp10:lss:pA)nu101;lssnu60 mutants (Fig. 5J,J′), col10a1a-positive chondrocytes were present in all growth plates studied (Fig. 5K,K′). These results indicate that although Lss activity is needed for proper growth plate development, loss of Msmo1 activity has a more-severe effect on chondrocyte differentiation. Importantly, the osteoblast expression of col10a1a in all mutants was similar to that in wild type (Fig. 5M-P), indicating a defect specific to chondrocyte differentiation. Interestingly, the domain of msmo1 expression in wild-type growth plates is localized to the transition zone from the columnar to hypertrophic chondrocytes (Fig. 5C,C′). This domain is absent in kolnu7 mutants (Fig. 5F,F′), consistent with the proposed regulatory nature of the mutation. In contrast, the msmo1 expression domain is expanded in the Tg(fabp10a:msmo1:pA)nu100;msmo1nu81 and Tg(fabp10:lss:pA)nu101;lssnu60 mutants (Fig. 5I,I′,L,L′), suggesting de-repression of the pathway. Based on these results, we conclude that, during normal development, msmo1 is transiently upregulated in pre-hypertrophic chondrocytes, and loss of the ability to synthesize cholesterol impedes chondrocyte hypertrophic differentiation.
Fig. 5.
Loss of Msmo1 and Lss activities disrupts growth plate patterning. (A-P) Expression domains of col2a1a (A,D,G,J), col10a1a (B,E,H,K,M-P) and msmo1 (C,F,I,L) were analyzed within growth plates of wild type (A-C), kolnu7 (D-F), Tg(fabp10a:msmo1:pA)nu100;msmo1nu81 (G-I) and Tg(fabp10:lss:pA)nu101;lssnu60 (J-L) using RNAscope in situ hybridization. (A) Strong col2a1a expression is observed in round and stacked chondrocytes in wild type, corresponding to presumed resting and proliferating chondrocytes. (B) Expression of col10a1a in wild type is observed complementary to col2a1a expression in presumed hypertrophic chondrocytes. The expression of col2a1a is expanded in kolnu7 (D) and Tg(fabp10a:msmo1:pA)nu100;msmo1nu81 (G) growth plates, with a near-complete loss of col10a1a expression in kolnu7 (E) and Tg(fabp10a:msmo1:pA)nu100;msmo1nu81 (H). Expression of col2a1a (J) and col10a1a (K) in Tg(fabp10:lss:pA)nu101;lssnu60 appears less affected when compared to kolnu7(D,E) and Tg(fabp10a:msmo1:pA)nu100;msmo1nu81 (G,H). Expression of msmo1 is detected primarily in pre-hypertrophic chondrocytes of wild type (C); msmo1 expression is undetectable in kolnu7 growth plates (F), but is upregulated in transgenically rescued msmo1nu81 (I) and lssnu60 (L) mutants. (M-P) Expression of col10a1a in osteoblasts is unchanged between wild type and mutants. Images represent paraffin sections of the pterotic at 2 months of age (∼15 mm SL). Boxed areas in A-L are shown at higher magnification in A′-L′. Scale bars: 100 μm. Tg*, Tg(fabp10:msmo1:pA)nu100; Tg**, Tg(fabp10:lss:pA)nu101. wt n=6, kolnu7 n=6, Tg(fabp10a:msmo1:pA)nu100;msmo1nu81 n=3, Tg(fabp10:lss:pA)nu101;lssnu60 n=3.
Loss of Msmo1 and Lss activity within growth plates does not result in loss of Ihh signaling
Ihh is a key ligand in cartilage and bone development. Ihh is synthesized by pre-hypertrophic chondrocytes and stimulates chondrocyte proliferation and hypertrophy, as well as osteoblast differentiation (Hammond and Schulte-Merker, 2009; St-Jacques et al., 1999; Vortkamp et al., 1996). Given the critical role of cholesterol in the post-translational Hedgehog (Hh) modification (Chen et al., 2011; Porter et al., 1995), we investigated what potential effect loss of Msmo1 and Lss activity may have on Ihh signaling within growth plates. Zebrafish have two Ihh paralogs, ihha and ihhb, with partially overlapping expression domains (Avaron et al., 2006). The receptor patched 1 (ptch1), a transcriptional target of Hh signaling, is expressed in chondrocytes and the perichondrium of endochondral bone (Felber et al., 2011; Hammond and Schulte-Merker, 2009; St-Jacques et al., 1999; Vortkamp et al., 1996). We analyzed expression of ihha, ihhb and ptch1 by RNAscope in situ hybridization using the same stages and anatomical location as described above. In wild-type fish, a broader ihha domain of expression in the hypertrophic chondrocytes zone and a more restricted ihhb domain of expression in mostly the pre-hypertrophic chondrocytes zone (Fig. 6A,B) was observed. At the same time, strong ptch1 expression was observed in the adjacent columnar zone and in individual hypertrophic chondrocytes (Fig. 6C). Expression of ihha and ihhb was still observed in growth plates of kolnu7, Tg(fabp10a:msmo1:pA)nu100;msmo1nu81 and Tg(fabp10:lss:pA)nu101;lssnu60 mutants (Fig. 6D,E,G,H,J,K). Importantly, ptch1 expression was also detected in all mutant growth plates (Fig. 6F,I,L), indicating that loss of cholesterol biosynthesis in the mutants does not abolish Ihh signaling activity.
Fig. 6.
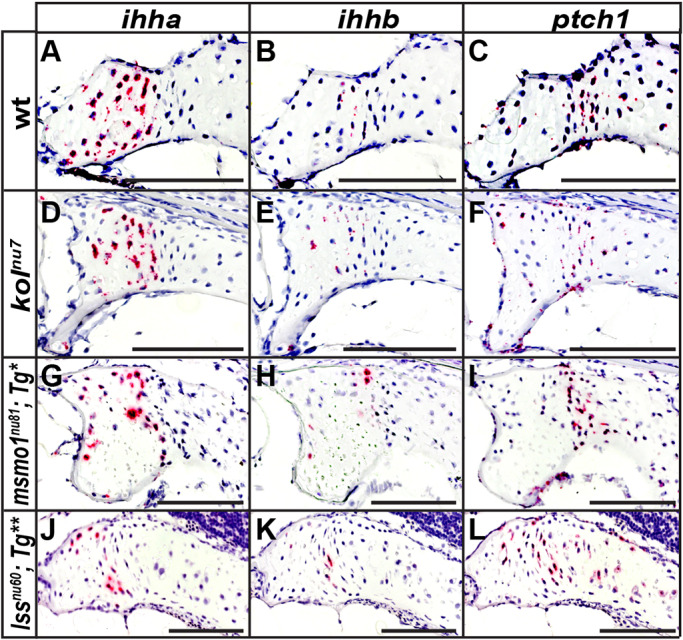
Ihh signaling within endochondral growth plates is present despite loss of Msmo1 and Lss activity. (A-L) Expression domains of ihha (A,D,G,J), ihhb (B,E,H,K) and ptch1 (C,F,I,L) were analyzed within growth plates of wild type (A-C), kolnu7 (D-F), Tg(fabp10a:msmo1:pA)nu100;msmo1nu81 (G-I) and Tg(fabp10:lss:pA)nu101;lssnu60 (J-L) using RNAscope in situ hybridization. (A-C) In wild type, expression of ihha and ihhb is observed in the pre-hypertrophic chondrocyte zone, with ptch1 expression seen to either side of these domains. (D-L) Expression of ihha, ihhb and ptch1 is present within mutant growth plates and similar to domains observed in wild type. Images represent paraffin sections of the pterotic at 2 months of age (∼15 mm SL). Scale bars: 100 μm. Tg*, Tg(fabp10:msmo1:pA)nu100; Tg**, Tg(fabp10:lss:pA)nu101. wt n=3, kolnu7 n=3, kolnu7/+:msmolnu81/+ n=3, Tg(fabp10a:msmo1:pA)nu100;msmo1nu81 n=3, Tg(fabp10:lss:pA)nu101;lssnu60 n=3.
DISCUSSION
Zebrafish post-squalene mutants show phenotypic similarities to human CDP
Many post-squalene pathway disorders with documented CDP result from partial loss-of-function mutations, such as mutations in the NAD(P)-dependent steroid dehydrogenase-like (NSDHL) gene (Fig. S1), which result in congenital hemidysplasia with ichthyosiform erythroderma and limb defects (CHILD) syndrome (Bornholdt et al., 2005; Mi et al., 2015; Herman and Kratz, 2012), and mutations in the emopamil binding protein (EBP) gene (Fig. S1), which result in X-linked dominant chondrodysplasia punctata (CDPX2), or Conradi-Hunermann-Happle syndrome (Cañueto et al., 2014; Herman and Kratz, 2012). Similarly, the kolnu7, kolnu7/+:msmo1nu81/+ and Tg(fabp10a:msmo1:pA)nu100;msmo1nu81 mutants result from reduction and/or absence of Msmo1 activity within skeletal elements and display ectopic ossification within growth plates similar to that seen in CDP (Figs 4 and 7) (Cañueto et al., 2014; Herman and Kratz, 2012; Rossi et al., 2015). As such, these mutants could prove helpful as animal models to help understand the mechanisms behind the development, and potential treatment, of CDP.
Fig. 7.
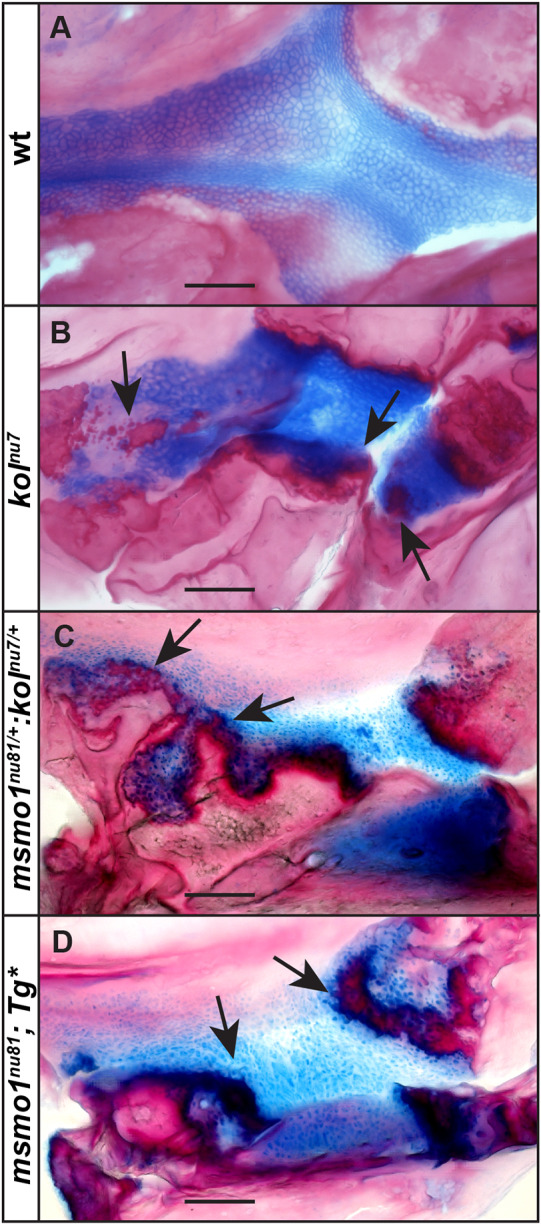
New zebrafish models for CDP. (A-D) Abnormal ossification, marked with arrows, within endochondral growth plates of kolnu7 (B), kolnu7/+:msmolnu81/+ (C) and Tg(fabp10a:msmo1:pA)nu100;msmo1nu81 (D) mutant phenotypes resembles defects observed in patients with chondrodysplasia punctata. Images represent lateral views of palatoquadrate growth plates of craniofacial bones from disarticulated whole-mount adult skeletal preparations at ∼4 months of age. Scale bars: 100 μm. Tg*, Tg(fabp10:msmo1:pA)nu100. wt n=4, kolnu7 n=6, kolnu7/+:msmolnu81/+ n=4, Tg(fabp10a:msmo1:pA)nu100;msmo1nu81 n=4.
Sterol intermediates may play a role in msmo1nu81 mutant phenotype
A lack of cholesterol alone does not explain all of the defects observed in post-squalene cholesterol biosynthesis disorders, as many patients present with normal cholesterol levels (Porter, 2008; Porter and Herman, 2011). Hypomorphic mutations in MSMO1 result in the rare autosomal recessive disorder C4 methyloxidase-like deficiency (He et al., 2011). Since patients may not present with decreased cholesterol levels, the definitive test for this disorder is an accumulation of C4 sterol intermediates (Frisso et al., 2017; He et al., 2011, 2014; Herman and Kratz, 2012; Rossi et al., 2015). Similarly, patients with Smith-Lemli-Opitz syndrome, which results from mutations in the 7-dehydrocholesterol reductase (DHCR7) gene (Fig. S1), and CDPX2, a result of mutations in the EBP gene (Fig. S1), often present with normal cholesterol levels and elevated levels of sterol intermediates (Braverman et al., 1999; Porter, 2008; Porter and Herman, 2011; Wassif et al., 1998).
Treatment regimens for many post-squalene pathway disorders may include dietary cholesterol supplementation and statin drug therapy (Porter and Herman, 2011). Statins act by impeding cholesterol synthesis through inhibition of HMG-CoA reductase (Sharpe and Brown, 2013) (Fig. S1). It is thought that statins may help prevent a buildup of potentially toxic sterol intermediates. Similar to statin treatment, inactivation of LSS activity impedes cholesterol synthesis and prevents sterol intermediate accumulation. But, unlike statin treatment, inactivation of LSS does not affect isoprenoid synthesis (Fig. S1). The protective effect provided by loss of Lss function in the msmo1nu81 mutants supports this prediction. Both msmo1nu81 and lssnu60 mutants are deficient in cholesterol biosynthesis, and both are characterized by progressive growth retardation and lethality soon after depletion of maternally deposited lipid and cholesterol-rich yolk (Miyares et al., 2014; Quinlivan and Farber, 2017) (Figs 2A and 3B). However, the lssnu60 mutants live 3 days longer (Fig. 3B) than msmo1nu81 mutants (Fig. 2A), suggesting that toxic sterol intermediates may contribute to the death of msmo1nu81 mutants. Importantly, the earlier lethality of the msmo1nu81 mutants can be suppressed by a loss of Lss activity (Fig. 3C), indicating a protective effect through inhibition of sterol intermediate formation.
Correspondingly, a more-severe patterning defect is observed in Tg(fabp10a:msmo1:pA)nu100;msmo1nu81 mutant growth plates compared to Tg(fabp10:lss:pA)nu101;lssnu60 mutants. In particular, the col10a1a expression in kolnu7 (Fig. 5E,E′) and Tg(fabp10a:msmo1:pA)nu100;msmo1nu81 (Fig. 5H,H′) mutant growth plates is nearly absent. At the same time, the col10a1a-positive domain is reduced but not completely eliminated in Tg(fabp10:lss:pA)nu101;lssnu60 mutants (Fig. 5K,K′). Our results suggest that both Msmo1 and Lss activity are needed for proper growth plate patterning and efficient chondrocyte differentiation. However, blockage of the pathway at the C4 demethylation step is associated with a more-severe defect, indicating a role for sterol intermediates, in addition to cholesterol depletion, in growth plate patterning and chondrocyte differentiation. Our findings point to Lss as an interesting target to mitigate some symptoms associated with disorders of the post-squalene cholesterol biosynthesis pathway without disrupting isoprenol-dependent processes.
Growth plates of kolnu7 and Tg(fabp10a:msmo1:pA)nu100;msmo1nu81 mutants show distinct phenotype
The growth plates of kolnu7 and Tg(fabp10a:msmo1:pA)nu100;msmo1nu81 are unique in that they display an enhanced and ectopic ossification phenotype (Fig. 4E-H,M-P), yet patterning of the growth plates revealed loss of col10a1a expression and absence of the presumptive hypertrophic chondrocyte zone (Fig. 5E,E′,H,H′). Interestingly, Ihh genes are still expressed in the mutants, suggesting that the transition from proliferating to pre-hypertrophic zone is not inhibited. The enhanced ossification phenotype seen in kolnu7 and Tg(fabp10a:msmo1:pA)nu100;msmo1nu81 mutants appears similar to that in zebrafish cyp26b1 mutants, but these mutants do not show a loss of col10a1a expression in endochondral bones (Laue et al., 2008; Spoorendonk et al., 2008). The unique phenotype of kolnu7 and Tg(fabp10a:msmo1:pA)nu100;msmo1nu81 mutants most closely resembles Sox9 inactivation in mouse growth plates (Dy et al., 2012; Ikegami et al., 2011). Sox9 is a master regulator of chondrocyte differentiation and survival (Lefebvre et al., 2019), indicating that Msmo1 activity within pre-hypertrophic chondrocytes might play a role in controlling cell hypertrophy and chondrocyte differentiation within the growth plate.
These results indicate that cholesterol depletion, and possibly accumulation of sterol intermediates, within mutant chondrocytes exiting from the proliferation zone may prevent normal differentiation and proper growth plate patterning, leading to an accelerated ossification. Intriguingly, CDP is observed in patients with a deficit in the initial steps of the post-squalene cholesterol biosynthesis pathway (Rossi et al., 2015), pointing to a potential active role of sterol intermediates in the first steps of biosynthesis in bone pathology. Recently, sterol intermediates were implicated in interactions with numerous signaling pathways by acting as ligands for LXRs (also known as NR1Hs), RORγ (also known as RORC) and EGFR (Gabitova et al., 2015; Hu et al., 2015; Santori et al., 2015; Sukhanova et al., 2013; Urlep et al., 2017; Yang et al., 2006). These data highlight the importance of sterol intermediates in the cholesterol biosynthesis pathway and the role these intermediates may play in skeletal dysplasia.
Msmo1 and Lss activity in growth plates is not essential for Ihh signaling
It is currently proposed that defects observed in disorders of the post-squalene pathway are due to two main mechanisms: abnormal Hh signaling and accumulation of sterol intermediates (Porter and Herman, 2011). Cholesterol modification of the Hh protein is necessary for proper Hh signaling and influences the spread of the morphogen through tissue (Briscoe and Thérond, 2013; Zeng, 2001; Zeng et al., 2001). Recently, it has been shown that cholesterol interacts with Smoothened (SMO) and the inability of SMO to bind cholesterol negatively affects Hh signaling (Byrne et al., 2016).
The role of Hh signaling in skeletogenesis is well known, with Shh playing a pivotal role in early patterning and Ihh acting later during bone formation (Yang et al., 2015). Thus, defects resulting in impaired cholesterol biosynthesis can affect Hh signaling, and many bone defects seen in cholesterol biosynthesis disorders, such as polydactyly, syndactyly and cleft palate, are hallmarks of abnormal Shh signaling (Briscoe and Thérond, 2013; Cooper et al., 2003; Gao et al., 2009; Hui, 1993; Hui and Joyner, 1993). It is interesting to note that although the kolnu7, msmo1nu81 and lssnu60 mutations negatively affect cholesterol synthesis, these mutants do not display the classical patterning defects associated with defective Shh signaling (Eberhart et al., 2006; Schwend and Ahlgren, 2009; Wada et al., 2005) (Fig. S4). This is likely to be caused by maternally deposited cholesterol within the yolk of the zebrafish that allows for normal embryonic and early larval development (Miyares et al., 2014; Quinlivan and Farber, 2017). Zebrafish have a large yolk that is slowly absorbed over the first 5 days of development (Miyares et al., 2014; Quinlivan and Farber, 2017). Therefore, zebrafish larvae do not require an endogenous biosynthesis of cholesterol until ∼6 dpf.
Although juvenile kolnu7 mutants display an absence of msmo1 expression in growth plates (Fig. 5F,F′), Ihh signaling within kolnu7 growth plates is not lost (Fig. 6D-F). Similarly, juvenile Tg(fabp10a:msmo1:pA)nu100;msmo1nu81 and Tg(fabp10:lss:pA)nu101;lssnu60 mutants exhibit loss of Msmo1 and Lss activity, respectively, yet expression of Ihh ligands and their transcriptional target ptch1 can be detected (Fig. 6G-L). These results suggest that the level of cholesterol in chondrocytes depleted of Msmo1 and Lss activity is sufficient to maintain Hh signaling.
The kolnu7 mutant highlights the significance of cis-acting regulatory mutations in disease
The importance of regulatory elements in human health and disease has been well recognized (Venter et al., 2001; Wray, 2007). With protein-coding exons making up less than 2% of the human genome (Venter et al., 2001), cis-regulatory elements are now believed to be more important than gene-encoding sequences in determining human traits and susceptibility to disease (Niu et al., 2018). In fact, many skeletal malformations are the result of mutations in regulatory elements that control expression of genes such as bone morphogenetic protein 2 (BMP2) (Dathe et al., 2009), SOX9 (Kurth et al., 2009), distal-less homeobox 5/6 (DLX5/6) (Kouwenhoven et al., 2010) and SHH (Albuisson et al., 2011; Wieczorek et al., 2010). Given the importance of regulatory elements in disease, the identification of new cis-regulatory mutations and the genes they regulate is crucial. The region harboring the kolnu7 locus and msmo1 gene is highly syntenic with a corresponding region of human chromosome 4 (data not shown), raising the possibility that similar long-distance regulation mechanisms operate in higher vertebrates.
Mutations in msmo1 and lss reveal an important role of cholesterol biosynthesis genes in bone development
Previously, very little was known of the role MSMO1 and LSS played in bone development. Of the five reported cases of C4 methyloxidase-like deficiency, symptoms reported include cataracts, intellectual delays, skin abnormalities, immune dysfunction, shortened stature and microcephaly (Frisso et al., 2017; He et al., 2011, 2014; Herman and Kratz, 2012; Rossi et al., 2015). Of the seven published articles on MSMO1 gene function, none show its role in bone development. Similarly, very few published articles show a link between LSS gene function and bone development. In humans, hypomorphic mutations in LSS result in congenital cataracts (Chen and Liu, 2017; Zhao et al., 2015), hypotrichosis simplex (Romano et al., 2018) and alopecia with mental retardation syndrome (Besnard et al., 2019).
Here, for the first time, we show that msmo1 and lss are needed for proper chondrocyte differentiation, growth plate formation and endochondral ossification. To our knowledge, the msmo1nu81 and lssnu60 mutants reported here are the first-described loss-of-function fish mutants in Msmo1 and Lss, respectively. The kolnu7 and msmo1nu81 mutants are the first-reported animal models for the human disorder C4 methyloxidase-like deficiency. These mutants, along with the stable transgenic mutant lines Tg(fabp10a:msmo1:pA)nu100;msmo1nu81 and Tg(fabp10:lss:pA)nu101;lssnu60 described above, provide essential tools to further elucidate the roles of msmo1 and lss in skeletal dysplasia. Zebrafish are an excellent tool for pre-clinical drug testing (MacRae and Peterson, 2015; Tan and Zon, 2011); thus, the lines presented here may provide invaluable insight into treatment options for those suffering from C4 methyloxidase-like deficiency, as well as other disorders of the post-squalene cholesterol biosynthesis pathway.
MATERIALS AND METHODS
Positional cloning
The kolnu7 mutation was localized to zebrafish chromosome 1 using a genetic linkage map of sequence-length polymorphism markers (Knapik et al., 1998). Additional closely linked markers, based on either restriction enzyme polymorphism or sequencing for single-nucleotide polymorphism were used to narrow down the kolnu7 location to a ∼457 kb critical region (Fig. 1C).
MultAlin analysis
All protein sequences were obtained from UniProtKB (UniProt, 2019): UGT8 Homo sapiens (Entry ID: Q16880), UGT8 Gallus gallus (Entry ID: F1NH08) and Ugt8 Danio rerio (Entry ID: Q1ECX6). These sequences were then aligned using MultAlin (Corpet, 1988), using an absolute scoring method and Blosum62 symbol comparison table with a gap opening default value of 12 and gap extension default value of 2.
RNA extraction
To extract bone-enriched total RNA, the hypural complexes from kolnu7 and wild-type siblings were dissected after the SL for each animal was recorded. Each hypural complex was placed in a 1.5-ml RINO Navy Bead tube (RINO BulletBlender Navy Bead Lysis kit; Next Advance, Troy, NY, USA) with 600 μl TRIzol (Invitrogen). Tissue was homogenized using the BeadBug Microtube Homogenizer (Benchmark Scientific, Sayreville, NJ, USA). Samples were extracted using five cycles of 1 min homogenization followed by 1 min cooling in a wet ice bath. RNA was purified from the TRIzol solution using Directzol RNA Minipreps (Zymo Research, Irvine, CA, USA).
Quantitative RT-PCR
All assays were performed using an EXPRESS One-Step Superscript qRT-PCR Kit, with premixed ROX (#11791200; Thermo Fisher Scientific, Waltham, MA, USA), Applied Biosystems MicroAmp Fast Optical reaction plates and 8-cap strips (Thermo Fisher Scientific) on an Applied Biosystems StepOnePlus Real-Time PCR System. For each reaction, 8 ng total RNA was used. All assays consisted of three technical replicates and a minimum of two biological replicates. The following Made to Order Taqman probes (Thermo Fisher Scientific) were used: mettl14 (Assay ID: Dr03438758_g1), tll1 (Assay ID: Dr03138096_m1), cpe (Assay ID: Dr03188311_s1), grhprb (Assay ID: Dr03088390_g1), ugt8 (Assay ID: Dr03427025) msmo1 (Assay ID: Dr03133463_m1) and eef1a1l1 (Assay ID: Dr03432748_m1). In addition, the following Custom Plus Taqman probes (Thermo Fisher Scientific) were designed and used: ndst3 (Assay ID: AJY9Y10), spock3 (Assay ID: AJ20TKF), prss12 (Assay ID: AJ5IPWS), apbb2 (Assay ID: AJT96CS) and uba6 (Assay ID: AJPADNW). The efficiency of each probe was tested prior to experimentation using the following equation: PCR efficiency (%)=(10(–1/S) – 1)×100, where S=slope of the standardized curve.
Semi-quantitative RT-PCR
All assays were performed using a Superscript III One-Step RT-PCR System with Platinum Taq DNA Polymerase. For each reaction, 30 ng RNA was used. The wild-type, kolnu7 and msmo1nu81 alleles were amplified using the primers 5′-TGGTCTTATGTGAACTTTCTTTACA-3′ and 5′- GATAAATCCAGGCAGGCAGA-3′. The PCR products were separated using a 3.5% Metaphor agarose (Lonza) gel. All assays consisted of three biological replicates.
CRISPR/Cas9 genome editing
Genome editing was performed as previously described (Hwang et al., 2013; Jao et al., 2013). We used linearized plasmid pCS2-nCas9n (Addgene plasmid #47929) (Jao et al., 2013) as a template for Cas9 mRNA synthesis (mMessage mMachine; Thermo Fisher Scientific). The guide RNA (gRNA) template was made by PCR and RNA was synthesized using a MEGAshortscript T7 kit (Thermo Fisher Scientific). In all cases, 30-40 pg gRNA was co-injected with 150 pg Cas9 mRNA per embryo at one-cell stage. Indels were identified by loss of restriction enzyme sites. The G0 founders positive for mutation were outcrossed with wild-type fish to produce F1 heterozygotes, which were then analyzed using sequencing to identify the mutations. Individuals carrying desirable, identified mutations were outcrossed with wild type to generate an F2 heterozygous family.
High-cholesterol diet (HCD)
Beginning at 5 dpf, larvae were fed a HCD similar to that previously described (Stoletov et al., 2009). Fish were raised in 450 ml system water, 6 parts per thousand (ppt) salinity, in 2.8-l tanks, and fed 15 mg of either a normal diet or HCD twice daily. Water was changed daily. Normal (0.12% cholesterol) and HCD (4% cholesterol) was provided by the Department of Biology, University of Alabama at Birmingham, Birmingham, AL, USA (Watts et al., 2012).
Total cholesterol measurement
Mutant clutches were collected in Petri dishes filled with egg water at 0 dpf. Embryos were placed in a 28°C incubator and fasted until collection at 8 dpf when they were snap frozen. An eyeball of each larva was removed for genotyping purposes. Once genotyping results were confirmed, individual larvae were homogenized in 200 µl extraction buffer chloroform:isopropanol:IGEPAL CA-630 (7:11:0.1) using a micro-tube homogenizer. The extraction procedure and total cholesterol measurements were carried out using a BioVision Total Cholesterol and Cholesteryl Ester Colorimetric/Fluorometric Assay Kit (#K603-100), as per the manufacturer’s recommendations, with 25 µl extracted larva sample used per assay, and 25 µl Reaction Mix or 25 µl Background Control Mix used per well. All assays consisted of three technical sample replicates, three technical background control replicates and three biological replicates. Fluorescence was detected using a CLARIOstar Microplate Reader (BMG Labtech, Ortenberg, Germany). Mean relative fluorescence units (RFUs) for each sample were calculated by subtracting the mean background control value from the mean sample value. From these data, the mean RFU for each group was calculated. Wild-type mean RFU=206081, s.d. 8150, s.e.m. 4705, n=3; msmo1nu81 mean RFU=45401, s.d. 26629, s.e.m. 15374, n=3 (P=0.0006); lssnu60 mean RFU=89447, s.d. 22756, s.e.m. 13138, n=3 (P=0.0011). Mutant RFU levels were compared to wild-type RFU levels to determine the percentage of total cholesterol levels. P-values were calculated using unpaired Student's t-test.
Embryonic cell transplantation
Transplantation was performed as previously described (Ho and Kane, 1990; Stafford et al., 2006). Wild-type donor embryos were co-injected at one- to two-cell stage with 20 ng/μl sox32 mRNA and 40 kD fluorescein dextran to allow for visualization of donor cells. Approximately 20-30 cells from high-stage donors [∼3 h post-fertilization (hpf)] were distributed along the blastoderm margin of shield-stage (∼6 hpf) hosts generated form a msmo1nu81 heterozygote in-cross. Then, 3 dpf embryos with a significant number of transplanted cells located in the intestines were selected for growth. At ∼6 weeks of age, fish were photographed using a Zeiss V-8 stereomicroscope equipped with a Zeiss Axiocam camera. To detect Tg(ubi:Zebrabow-M)a131 donor cells, a Texas Red filter was used. Host genotype was determined based on DNA extracted from a fin clip devoid of red, transplanted cells.
Creation of transgenic lines
The genomic and coding sequences of msmo1 and lss were obtained from Wellcome Trust Sanger Ensembl database, Danio rerio (GRCz11). The open-reading frame (ORF) of msmo1 was amplified from wild-type RNA; the ORF of lss was amplified from cDNA clone (NM_001083567.1) and cloned using Gateway BP reaction on the pDONOR221 plasmid (Invitrogen). The fabp10 promoter was amplified from pHD157 (Addgene plasmid #84033) (Ni et al., 2012) using primers 5′-CTGAGCATCAGAATGGGGAAG-3′ and 5′-GCTCAACACAAAGTGAAGGTC-3′, and re-amplified with primers 5′-TGATATCGAATTCCTGCAGCCTGAGCATCAGAATGGGG-3′ and 5′-CGCTCTAGAACTAGTGGATCGCTCAACACAAAGTGAAG-3′ to direct proper orientation of the insert. Using NEBuilder HiFi DNA Assembly Master Mix, this product was then subcloned into the p5E-MCS (Addgene plasmid #26029) (Quillien et al., 2017) linearized with SmaI. Finally, plasmids were assembled using Gateway LR reaction and clones available in the Tol2kit (Kwan et al., 2007). To create the hsp70l:msmo1:IRESnlsEGFP plasmid, the ORF of msmo1 was cloned between the hsp70l promoter and the IRESnlsEGFP sequence in the pDestTol2CG2 vector. To create the fabp10:msmo1:pA and fabp10:lss:pA lines, the ORF of msmo1 or lss was cloned between the fabp10 promoter and a polyA sequence in the pDestTol2CG2 vector. All transgenic lines were generated with the help of the Tol2 transposase method of transgenesis (Kawakami and Shima, 1999; Kwan et al., 2007). As the transgene backbone contains a cmlc2:EGFP element, all transgene-positive fish were identified at 2 dpf by EGFP-positive hearts.
Heat-shock protocol
To induce transgene expression in Tg(hsp70l:msmo1:IRESnlsEGFP)nu99 progeny of the identified msmo1nu81 heterozygote crosses, fish were heat shocked daily at 42°C for 15 min, beginning at 24 hpf until collection and genotyping at 14 dpf.
Histological sections
Two-month old fish were fixed overnight in phosphate-buffered 10% formalin and treated for 8 h before processing with 0.5 M EDTA, pH 8. Samples were processed using an STP 120 Spin Tissue Processor (Thermo Fisher Scientific). Dehydration steps were conducted gradually with 10%, 30%, 50%, and 70% ethanol, each for 1 h. Final ethanol treatments of 95% and 100% were each performed twice, both for 1 h each. Xylene was applied twice, 45 min each time, and paraffin (Histoplast Paraffin LP 8332; Thermo Fisher Scientific) applied three times for 60 min each time. Specimens were embedded using a Microm EC350-2 tissue-embedding center. All tissue samples were sectioned at 5 μm using a rotary microtome.
In situ hybridization
The 918 bp template for the msmo1 anti-sense RNA probe was amplified from wild-type RNA using a primer tailed with the T7 RNA polymerase promoter (5′-TGGTCTTATGTGAACTTTCTTTACA-3′ and 5′-TAATACGACTCACTATAGGGGTCTGTCCCACCAGGTGAAT-3′). Whole-mount in situ hybridization was performed as previously described (Thisse and Thisse, 2008), using low-stringency conditions. Hybridization mix was supplemented with 5% dextran sulfate to increase signal intensity (Lauter et al., 2011; Thisse and Thisse, 2014). RNAscope in situ hybridization was carried out on formalin-fixed, paraffin-embedded (FFPE) 5-μm sections. All RNA probes, hybridization kits and hybridization equipment, including ACD HybEZ Hybridization oven, were obtained from Advanced Cell Diagnostics (Newark, CA, USA). All FFPE sections were treated with an RNAscope 2.5 Universal Pretreatment Reagents kit and RNAscope 2.5 High Definition (HD)-Red Assay kit, as per the manufacturer’s recommendations, with the following adjustments to the tissue pre-treatment protocol. Slides were heated to 98-102°C in 1× Target Retrieval Reagents for 8 min and incubated at 40°C in Protease Plus for 15 min. Staining intensity for each probe was adjusted by varying the incubation time of Hybridization AMP 5. Signal detection time for each probe was as follows: 5 min for col2a1a (#409471) and col10a1a (#409481) or 30 min for msmo1 (#438981), ihha (#490681), ihhb (#490691) and ptch1 (#490701). The probe dapB (#310043) was included as a negative control in each assay, and incubated for either 5 min or 30 min depending on the assay. Each probe was tested in a minimum of three independent experiments on multiple sections. Slides were counterstained with 50% Hematoxylin and 0.02% (w/v) ammonia water, as per the manufacturer's suggestions. Photographs were taken using a Zeiss Axioplan 2 stereomicroscope equipped with a Zeiss Axiocam camera.
Bone and cartilage staining
Larvae and adult fish were euthanized by submersion in ice-cold system water and fixed in phosphate-buffered 10% formalin for 2 h (larvae) or 1-2 days (adults). After rinsing in PBS, specimens were dehydrated gradually to 70% ethanol for long-term storage at 4°C. Whole-mount skeletal stains for adult bone and cartilage were performed as previously described (Taylor and Vandyke, 1985). Bone-only or cartilage-only protocols were performed for larval fish as previously described (Detrich et al., 2004), with the following modification: larvae were stained in a 0.005% solution of Alizarin Red dissolved in 1% KOH overnight. When not in use, specimens were kept in the dark to avoid fading. Photographs were taken using Zeiss Stemi 2000-CS and Zeiss Axioplan 2 stereomicroscopes, both equipped with a Zeiss Axiocam camera.
Genotyping
For all genotyping protocols, small DNA fragments were amplified using the primers listed in Table S1. The kolnu7 genotyping protocol was developed based on the polymorphism in linked exon 5 of ndst3 that eliminates a SacI restriction enzyme site. The genotyping protocols for ndst3nu20, ugt8nu82 and spock3nu83 were based on the elimination of restriction enzyme sites (Table 1). The genotyping of msmo1nu81 and lssnu60 alleles takes advantage of a 37 bp insertion and a 23 bp deletion, respectively. The PCR products were separated using either a 3.5% Metaphor agarose (Lonza) gel (kolnu7, spock3nu83, msmo1nu81, lssnu60) or 2.0% agarose gel (ndst3nu20 and ugt8nu82).
Statistical analysis
All statistical calculations were carried out with GraphPad Prism using the Chi-square or t-test calculator as noted.
Zebrafish lines and maintenance
All fish used in this study were raised and cared for in accordance with an approved protocol by Ann and Robert H. Lurie Children's Hospital of Chicago Institutional Animal Care and Use Committee (IACUC; #13-036), and complied with National Institutes of Health standards provided in the ‘Guide for the Care and Use of Laboratory Animals’. The following wild-type, mutant and transgenic fish lines were used: AB (ZFIN ID: ZDB-GENO-960809-7), TU (ZFIN ID: ZDB-GENO-990623-3), Tg(ubi:Zebrabow-M)a131 (ZFIN ID: ZDB-ALT-130816-2) (Pan et al., 2013), kolnu7, msmo1nu81, lssnu60, ndst3nu20, ugt8nu82, spock3nu83, Tg(hsp70l:msmo1:IRESnlsEGFP)nu99, Tg(fabp10:msmo1:pA)nu100 and Tg(fabp10:lss:pA)nu101. All mutant fish described in the experiments are the results of crosses between heterozygous parents. The kolnu7 mutant line is the result of an uncharacterized, naturally occurring mutation.
Fish harvesting
Fish were collected at various developmental stages and SL was measured from snout to the end of the hypuralia, with any post-fixation adjustments applied to SL as necessary (Parichy et al., 2009).
Supplementary Material
Acknowledgements
We acknowledge the Stanley Manne Children's Research Institute Microscopy and Histology Group.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: R.A.A., J.T.; Methodology: R.A.A.; Validation: R.A.A., J.T.; Formal analysis: R.A.A.; Investigation: R.A.A., K.T.S., S.R.M.; Resources: R.A.A., E.E.L., J.M.T., J.T.; Data curation: R.A.A.; Writing - original draft: R.A.A.; Writing - review & editing: R.A.A., E.E.L., J.T.; Visualization: R.A.A.; Supervision: E.E.L., J.M.T., J.T.; Project administration: J.T.; Funding acquisition: R.A.A., J.T.
Funding
This work was supported by the National Institute of Dental and Craniofacial Research (F31DE023481-03 to R.A.A.), Children's Research Fund (OGS Award to R.A.A.), Lurie Children's Hospital (to J.T.) and the Walter S. and Lucienne Driskill Foundation (to J.T.).
Supplementary information
Supplementary information available online at http://dmm.biologists.org/lookup/doi/10.1242/dmm.042549.supplemental
References
- Ačimovič J., Goyal S., Košir R., Goličnik M., Perše M., Belič A., Urlep Z., Guengerich F. P. and Rozman D. (2016). Cytochrome P450 metabolism of the post-lanosterol intermediates explains enigmas of cholesterol synthesis. Sci. Rep. 6, 28462 10.1038/srep28462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albuisson J., Isidor B., Giraud M., Pichon O., Marsaud T., David A., Le Caignec C. and Bezieau S. (2011). Identification of two novel mutations in Shh long-range regulator associated with familial pre-axial polydactyly. Clin. Genet. 79, 371-377. 10.1111/j.1399-0004.2010.01465.x [DOI] [PubMed] [Google Scholar]
- Avaron F., Hoffman L., Guay D. and Akimenko M. A. (2006). Characterization of two new zebrafish members of the hedgehog family: atypical expression of a zebrafish indian hedgehog gene in skeletal elements of both endochondral and dermal origins. Dev. Dyn. 235, 478-489. 10.1002/dvdy.20619 [DOI] [PubMed] [Google Scholar]
- Besnard T., Sloboda N., Goldenberg A., Kury S., Cogne B., Breheret F., Trochu E., Conrad S., Vincent M., Deb W. et al. (2019). Biallelic pathogenic variants in the lanosterol synthase gene LSS involved in the cholesterol biosynthesis cause alopecia with intellectual disability, a rare recessive neuroectodermal syndrome. Genet. Med. 21, 2025 10.1038/s41436-019-0445-x [DOI] [PubMed] [Google Scholar]
- Bird N. C. and Mabee P. M. (2003). Developmental morphology of the axial skeleton of the zebrafish, Danio rerio (Ostariophysi: Cyprinidae). Dev. Dyn. 228, 337-357. 10.1002/dvdy.10387 [DOI] [PubMed] [Google Scholar]
- Bornholdt D., Konig A., Happle R., Leveleki L., Bittar M., Danarti R., Vahlquist A., Tilgen W., Reinhold U., Poiares Baptista A. et al. (2005). Mutational spectrum of NSDHL in CHILD syndrome. J. Med. Genet. 42, e17. 10.1136/jmg.2004.024448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braverman N., Lin P., Moebius F. F., Obie C., Moser A., Glossmann H., Wilcox W. R., Rimoin D. L., Smith M., Kratz L. et al. (1999). Mutations in the gene encoding 3 beta-hydroxysteroid-delta 8, delta 7-isomerase cause X-linked dominant Conradi-Hunermann syndrome. Nat. Genet.22, 291-294. 10.1038/10357 [DOI] [PubMed] [Google Scholar]
- Briscoe J. and Thérond P. P. (2013). The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 14, 416-429. 10.1038/nrm3598 [DOI] [PubMed] [Google Scholar]
- Byrne E. F., Sircar R., Miller P. S., Hedger G., Luchetti G., Nachtergaele S., Tully M. D., Mydock-McGrane L., Covey D. F., Rambo R. P. et al. (2016). Structural basis of Smoothened regulation by its extracellular domains. Nature 535, 517-522. 10.1038/nature18934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañueto J., Girós M. and Gonzalez-Sarmiento R. (2014). The role of the abnormalities in the distal pathway of cholesterol biosynthesis in the Conradi-Hunermann-Happle syndrome. Biochim. Biophys. Acta 1841, 336-344. 10.1016/j.bbalip.2013.09.002 [DOI] [PubMed] [Google Scholar]
- Chen X. and Liu L. (2017). Congenital cataract with LSS gene mutations: a new case report. J. Pediatr. Endocrinol. Metab. 30, 1231-1235. 10.1515/jpem-2017-0101 [DOI] [PubMed] [Google Scholar]
- Chen X., Tukachinsky H., Huang C.-H., Jao C., Chu Y.-R., Tang H.-Y., Mueller B., Schulman S., Rapoport T. A. and Salic A. (2011). Processing and turnover of the Hedgehog protein in the endoplasmic reticulum. J. Cell Biol. 192, 825-838. 10.1083/jcb.201008090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper M. K., Wassif C. A., Krakowiak P. A., Taipale J., Gong R., Kelley R. I., Porter F. D. and Beachy P. A. (2003). A defective response to Hedgehog signaling in disorders of cholesterol biosynthesis. Nat. Genet. 33, 508-513. 10.1038/ng1134 [DOI] [PubMed] [Google Scholar]
- Corpet F. (1988). Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16, 10881–10890. 10.1093/nar/16.22.10881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubbage C. C. and Mabee P. M. (1996). Development of the cranium and paired fins in the zebrafish danio rerio (Ostariophysi, Cyprinidae). J. Morphol. 229, 121-160. [DOI] [PubMed] [Google Scholar]
- Dale R. M. and Topczewski J. (2011). Identification of an evolutionarily conserved regulatory element of the zebrafish col2a1a gene. Dev. Biol. 357, 518-531. 10.1016/j.ydbio.2011.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dathe K., Kjaer K. W., Brehm A., Meinecke P., Nurnberg P., Neto J. C., Brunoni D., Tommerup N., Ott C. E., Klopocki E. et al. (2009). Duplications involving a conserved regulatory element downstream of BMP2 are associated with brachydactyly type A2. Am. J. Hum. Genet. 84, 483-492. 10.1016/j.ajhg.2009.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detrich H. W., Westerfield M. and Zon L. I. (2004). The Zebrafish: 2nd Edition Cellular and Developmental Biology. 2 edn: Academic Press. [Google Scholar]
- Dy P., Wang W., Bhattaram P., Wang Q., Wang L., Ballock R. T. and Lefebvre V. (2012). Sox9 directs hypertrophic maturation and blocks osteoblast differentiation of growth plate chondrocytes. Dev. Cell 22, 597-609. 10.1016/j.devcel.2011.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eames B. F., Amores A., Yan Y.-L. and Postlethwait J. H. (2012). Evolution of the osteoblast: skeletogenesis in gar and zebrafish. BMC Evol. Biol. 12, 27 10.1186/1471-2148-12-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhart J. K., Swartz M. E., Crump J. G. and Kimmel C. B. (2006). Early Hedgehog signaling from neural to oral epithelium organizes anterior craniofacial development. Development 133, 1069-1077. 10.1242/dev.02281 [DOI] [PubMed] [Google Scholar]
- Felber K., Croucher P. and Roehl H. H. (2011). Hedgehog signalling is required for perichondral osteoblast differentiation in zebrafish. Mech. Dev. 128, 141-152. 10.1016/j.mod.2010.11.006 [DOI] [PubMed] [Google Scholar]
- Frisso G., Gelzo M., Procopio E., Sica C., Lenza M. P., Dello Russo A., Donati M. A., Salvatore F. and Corso G. (2017). A rare case of sterol-C4-methyl oxidase deficiency in a young Italian male: Biochemical and molecular characterization. Mol. Genet. Metab. 121, 329-335. 10.1016/j.ymgme.2017.06.013 [DOI] [PubMed] [Google Scholar]
- Gabitova L., Restifo D., Gorin A., Manocha K., Handorf E., Yang D.-H., Cai K. Q., Klein-Szanto A. J., Cunningham D., Kratz L. E. et al. (2015). Endogenous sterol metabolites regulate growth of EGFR/KRAS-dependent tumors via LXR. Cell Rep 12, 1927-1938. 10.1016/j.celrep.2015.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B., Hu J., Stricker S., Cheung M., Ma G., Law K. F., Witte F., Briscoe J., Mundlos S., He L. et al. (2009). A mutation in Ihh that causes digit abnormalities alters its signalling capacity and range. Nature 458, 1196-1200. 10.1038/nature07862 [DOI] [PubMed] [Google Scholar]
- Hammond C. L. and Schulte-Merker S. (2009). Two populations of endochondral osteoblasts with differential sensitivity to Hedgehog signalling. Development 136, 3991-4000. 10.1242/dev.042150 [DOI] [PubMed] [Google Scholar]
- He M., Kratz L. E., Michel J. J., Vallejo A. N., Ferris L., Kelley R. I., Hoover J. J., Jukic D., Gibson K. M., Wolfe L. A. et al. (2011). Mutations in the human SC4MOL gene encoding a methyl sterol oxidase cause psoriasiform dermatitis, microcephaly, and developmental delay. J. Clin. Invest. 121, 976-984. 10.1172/JCI42650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M., Smith L. D., Chang R., Li X. and Vockley J. (2014). The role of sterol-C4-methyl oxidase in epidermal biology. Biochim. Biophys. Acta 1841, 331-335. 10.1016/j.bbalip.2013.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Her G. M., Yeh Y.-H. and Wu J.-L. (2003). 435-bp liver regulatory sequence in the liver fatty acid binding protein (L-FABP) gene is sufficient to modulate liver regional expression in transgenic zebrafish. Dev. Dyn. 227, 347-356. 10.1002/dvdy.10324 [DOI] [PubMed] [Google Scholar]
- Herman G. E. and Kratz L. (2012). Disorders of sterol synthesis: beyond Smith-Lemli-Opitz syndrome. Am. J. Med. Genet. C Semin. Med. Genet. 160C, 301-321. 10.1002/ajmg.c.31340 [DOI] [PubMed] [Google Scholar]
- Ho R. K. and Kane D. A. (1990). Cell-autonomous action of zebrafish spt-1 mutation in specific mesodermal precursors. Nature 348, 728-730. 10.1038/348728a0 [DOI] [PubMed] [Google Scholar]
- Hu X., Wang Y., Hao L.-Y., Liu X., Lesch C. A., Sanchez B. M., Wendling J. M., Morgan R. W., Aicher T. D., Carter L. L. et al. (2015). Sterol metabolism controls T(H)17 differentiation by generating endogenous RORgamma agonists. Nat. Chem. Biol. 11, 141-147. 10.1038/nchembio.1714 [DOI] [PubMed] [Google Scholar]
- Huff M. W. and Telford D. E. (2005). Lord of the rings--the mechanism for oxidosqualene:lanosterol cyclase becomes crystal clear. Trends Pharmacol. Sci. 26, 335-340. 10.1016/j.tips.2005.05.004 [DOI] [PubMed] [Google Scholar]
- Hui C. (1993). A mouse model of Greig cephalopolysyndactyly syndrome: the extra toesJ mutation contains an intragenic deletion of the Gli3 gene. Nature 3, 241-246. 10.1038/ng0393-241 [DOI] [PubMed] [Google Scholar]
- Hui C.-C. and Joyner A. L. (1993). A mouse model of greig cephalopolysyndactyly syndrome: the extra-toesJ mutation contains an intragenic deletion of the Gli3 gene. Nat. Genet. 3, 241-246. 10.1038/ng0393-241 [DOI] [PubMed] [Google Scholar]
- Hwang W. Y., Fu Y., Reyon D., Maeder M. L., Tsai S. Q., Sander J. D., Peterson R. T., Yeh J.-R. J. and Joung J. K. (2013). Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31, 227-229. 10.1038/nbt.2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami D., Akiyama H., Suzuki A., Nakamura T., Nakano T., Yoshikawa H. and Tsumaki N. (2011). Sox9 sustains chondrocyte survival and hypertrophy in part through Pik3ca-Akt pathways. Development 138, 1507-1519. 10.1242/dev.057802 [DOI] [PubMed] [Google Scholar]
- Irving M. D., Chitty L. S., Mansour S. and Hall C. M. (2008). Chondrodysplasia punctata: a clinical diagnostic and radiological review. Clin. Dysmorphol. 17, 229-241. 10.1097/MCD.0b013e3282fdcc70 [DOI] [PubMed] [Google Scholar]
- Jao L.-E., Wente S. R. and Chen W. (2013). Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. USA 110, 13904-13909. 10.1073/pnas.1308335110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jira P. (2013). Cholesterol metabolism deficiency. Handb. Clin. Neurol. 113, 1845-1850. 10.1016/B978-0-444-59565-2.00054-X [DOI] [PubMed] [Google Scholar]
- Jurkiewicz E., Marcinska B., Bothur-Nowacka J. and Dobrzanska A. (2013). Clinical and radiological pictures of two newborn babies with manifestations of chondrodysplasia punctata and review of available literature. Pol. J. Radiol. 78, 57-64. 10.12659/PJR.883947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K. and Shima A. (1999). Identification of the Tol2 transposase of the medaka fish Oryzias latipes that catalyzes excision of a nonautonomous Tol2 element in zebrafish Danio rerio. Gene 240, 239-244. 10.1016/S0378-1119(99)00444-8 [DOI] [PubMed] [Google Scholar]
- Kim Y.-I., Lee S., Jung S.-H., Kim H.-T., Choi J. H., Lee M.-S., You K.-H., Yeo S.-Y., Yoo K.-W., Kwak S. A. et al. (2013). Establishment of a bone-specific col10a1:GFP transgenic zebrafish. Mol. Cells 36, 145-150. 10.1007/s10059-013-0117-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapik E. W., Goodman A., Ekker M., Chevrette M., Delgado J., Neuhauss S., Shimoda N., Driever W., Fishman M. C. and Jacob H. J. (1998). A microsatellite genetic linkage map for zebrafish (Danio rerio). Nat. Genet. 18, 338-343. 10.1038/ng0498-338 [DOI] [PubMed] [Google Scholar]
- Kouwenhoven E. N., van Heeringen S. J., Tena J. J., Oti M., Dutilh B. E., Alonso M. E., de la Calle-Mustienes E., Smeenk L., Rinne T., Parsaulian L. et al. (2010). Genome-wide profiling of p63 DNA-binding sites identifies an element that regulates gene expression during limb development in the 7q21 SHFM1 locus. PLoS Genet. 6, e1001065 10.1371/journal.pgen.1001065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg H. M. (2003). Developmental regulation of the growth plate. Nature 423, 332-336. 10.1038/nature01657 [DOI] [PubMed] [Google Scholar]
- Kurth I., Klopocki E., Stricker S., van Oosterwijk J., Vanek S., Altmann J., Santos H. G., van Harssel J. J., de Ravel T., Wilkie A. O. et al. (2009). Duplications of noncoding elements 5′ of SOX9 are associated with brachydactyly-anonychia. Nat. Genet. 41, 862-863. 10.1038/ng0809-862 [DOI] [PubMed] [Google Scholar]
- Kwan K. M., Fujimoto E., Grabher C., Mangum B. D., Hardy M. E., Campbell D. S., Parant J. M., Yost H. J., Kanki J. P. and Chien C. B. (2007). The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088-3099. 10.1002/dvdy.21343 [DOI] [PubMed] [Google Scholar]
- Laue K., Janicke M., Plaster N., Sonntag C. and Hammerschmidt M. (2008). Restriction of retinoic acid activity by Cyp26b1 is required for proper timing and patterning of osteogenesis during zebrafish development. Development 135, 3775-3787. 10.1242/dev.021238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauter G., Söll I. and Hauptmann G. (2011). Two-color fluorescent in situ hybridization in the embryonic zebrafish brain using differential detection systems. BMC Dev. Biol. 11, 43 10.1186/1471-213X-11-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClair E. E., Mui S. R., Huang A., Topczewska J. M. and Topczewski J. (2009). Craniofacial skeletal defects of adult zebrafish Glypican 4 (knypek) mutants. Dev. Dyn. 238, 2550-2563. 10.1002/dvdy.22086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V., Angelozzi M. and Haseeb A. (2019). SOX9 in cartilage development and disease. Curr. Opin. Cell Biol. 61, 39-47. 10.1016/j.ceb.2019.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Korzh V. and Gong Z. (2007). Localized rbp4 expression in the yolk syncytial layer plays a role in yolk cell extension and early liver development. BMC Dev. Biol. 7, 117 10.1186/1471-213X-7-117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. and Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lykissas M. (2013). Challenges of spine surgery in patients with chondrodysplasia punctata. J. Pediatr. Orthop. 33, 685-693. 10.1097/BPO.0b013e31829e86a9 [DOI] [PubMed] [Google Scholar]
- MacRae C. A. and Peterson R. T. (2015). Zebrafish as tools for drug discovery. Nat. Rev. Drug Discov. 14, 721-731. 10.1038/nrd4627 [DOI] [PubMed] [Google Scholar]
- Mi X. B., Luo M. X., Guo L. L., Zhang T. D. and Qiu X. W. (2015). CHILD syndrome: case report of a Chinese patient and literature review of the NAD[P]H steroid dehydrogenase-like protein gene mutation. Pediatr. Dermatol. 32, e277–e282. 10.1111/pde.12701 [DOI] [PubMed] [Google Scholar]
- Miyares R. L., de Rezende V. B. and Farber S. A. (2014). Zebrafish yolk lipid processing: a tractable tool for the study of vertebrate lipid transport and metabolism. Dis. Model. Mech. 7, 915-927. 10.1242/dmm.015800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudumana S. P., Wan H., Singh M., Korzh V. and Gong Z. (2004). Expression analyses of zebrafish transferrin, ifabp, and elastaseB mRNAs as differentiation markers for the three major endodermal organs: liver, intestine, and exocrine pancreas. Dev. Dyn. 230, 165-173. 10.1002/dvdy.20032 [DOI] [PubMed] [Google Scholar]
- Ni T. T., Lu J., Zhu M., Maddison L. A., Boyd K. L., Huskey L., Ju B., Hesselson D., Zhong T. P., Page-McCaw P. S. et al. (2012). Conditional control of gene function by an invertible gene trap in zebrafish. Proc. Natl. Acad. Sci. USA 109, 15389-15394. 10.1073/pnas.1206131109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu M., Tabari E., Ni P. and Su Z. (2018). Towards a map of cis-regulatory sequences in the human genome. Nucleic Acids Res. 46, 5395-5409. 10.1093/nar/gky338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz D. M. and Marie P. J. (2002). FGF signaling pathways in endochondral and intramembranous bone development and human genetic disease. Genes Dev. 16, 1446-1465. 10.1101/gad.990702 [DOI] [PubMed] [Google Scholar]
- Pan Y. A., Freundlich T., Weissman T. A., Schoppik D., Wang X. C., Zimmerman S., Ciruna B., Sanes J. R., Lichtman J. W. and Schier A. F. (2013). Zebrabow: multispectral cell labeling for cell tracing and lineage analysis in zebrafish. Development 140, 2835-2846. 10.1242/dev.094631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parichy D. M., Elizondo M. R., Mills M. G., Gordon T. N. and Engeszer R. E. (2009). Normal table of postembryonic zebrafish development: staging by externally visible anatomy of the living fish. Dev. Dyn. 238, 2975-3015. 10.1002/dvdy.22113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogoda H. M., Riedl-Quinkertz I., Lohr H., Waxman J. S., Dale R. M., Topczewski J., Schulte-Merker S. and Hammerschmidt M. (2018). Direct activation of chordoblasts by retinoic acid is required for segmented centra mineralization during zebrafish spine development. Development 145, dev159418 10.1242/dev.159418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter F. D. (2008). Smith-Lemli-Opitz syndrome: pathogenesis, diagnosis and management. Eur. J. Hum. Genet. 16, 535–541. 10.1038/ejhg.2008.10 [DOI] [PubMed] [Google Scholar]
- Porter F. D. and Herman G. E. (2011). Malformation syndromes caused by disorders of cholesterol synthesis. J. Lipid Res. 52, 6-34. 10.1194/jlr.R009548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter J. A., von Kessler D. P., Ekker S. C., Young K. E., Lee J. J., Moses K. and Beachy P. A. (1995). The product of hedgehog autoproteolytic cleavage active in local and long-range signalling. Nature 374, 363-366. 10.1038/374363a0 [DOI] [PubMed] [Google Scholar]
- Quillien A., Abdalla M., Yu J., Ou J., Zhu L. J. and Lawson N. D. (2017). Robust identification of developmentally active endothelial enhancers in zebrafish using FANS-assisted ATAC-Seq. Cell Rep 20, 709-720. 10.1016/j.celrep.2017.06.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlivan V. H. and Farber S. A. (2017). Lipid uptake, metabolism, and transport in the larval zebrafish. Front. Endocrinol. (Lausanne) 8, 319 10.3389/fendo.2017.00319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano M. T., Tafazzoli A., Mattern M., Sivalingam S., Wolf S., Rupp A., Thiele H., Altmuller J., Nurnberg P., Ellwanger J. et al. (2018). Bi-allelic mutations in LSS, encoding lanosterol synthase, cause autosomal-recessive hypotrichosis simplex. Am. J. Hum. Genet. 103, 777-785. 10.1016/j.ajhg.2018.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi M., Hall C. M., Bouvier R., Collardeau-Frachon S., Le Breton F., Bucourt M., Cordier M. P., Vianey-Saban C., Parenti G., Andria G. et al. (2015). Radiographic features of the skeleton in disorders of post-squalene cholesterol biosynthesis. Pediatr. Radiol. 45, 965-976. 10.1007/s00247-014-3257-9 [DOI] [PubMed] [Google Scholar]
- Santori F. R., Huang P., van de Pavert S. A., Douglass E. F. Jr., Leaver D. J., Haubrich B. A., Keber R., Lorbek G., Konijn T., Rosales B. N. et al. (2015). Identification of natural RORgamma ligands that regulate the development of lymphoid cells. Cell Metab. 21, 286-297. 10.1016/j.cmet.2015.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwend T. and Ahlgren S. C. (2009). Zebrafish con/disp1 reveals multiple spatiotemporal requirements for Hedgehog-signaling in craniofacial development. BMC Dev. Biol. 9, 59 10.1186/1471-213X-9-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe L. J. and Brown A. J. (2013). Controlling cholesterol synthesis beyond 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR). J. Biol. Chem. 288, 18707-18715. 10.1074/jbc.R113.479808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoorendonk K. M., Peterson-Maduro J., Renn J., Trowe T., Kranenbarg S., Winkler C. and Schulte-Merker S. (2008). Retinoic acid and Cyp26b1 are critical regulators of osteogenesis in the axial skeleton. Development 135, 3765-3774. 10.1242/dev.024034 [DOI] [PubMed] [Google Scholar]
- St-Jacques B., Hammerschmidt M. and McMahon A. P. (1999). Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 13, 2072-2086. 10.1101/gad.13.16.2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford D., White R. J., Kinkel M. D., Linville A., Schilling T. F. and Prince V. E. (2006). Retinoids signal directly to zebrafish endoderm to specify insulin-expressing beta-cells. Development 133, 949-956. 10.1242/dev.02263 [DOI] [PubMed] [Google Scholar]
- Stoletov K., Fang L., Choi S. H., Hartvigsen K., Hansen L. F., Hall C., Pattison J., Juliano J., Miller E. R., Almazan F. et al. (2009). Vascular lipid accumulation, lipoprotein oxidation, and macrophage lipid uptake in hypercholesterolemic zebrafish. Circ. Res. 104, 952-960. 10.1161/CIRCRESAHA.108.189803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukhanova A., Gorin A., Serebriiskii I. G., Gabitova L., Zheng H., Restifo D., Egleston B. L., Cunningham D., Bagnyukova T., Liu H. et al. (2013). Targeting C4-demethylating genes in the cholesterol pathway sensitizes cancer cells to EGF receptor inhibitors via increased EGF receptor degradation. Cancer Discov 3, 96-111. 10.1158/2159-8290.CD-12-0031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot W. S. and Schier A. F. (1999). Positional cloning of mutated zebrafish genes. Methods Cell Biol. 60, 259-286. 10.1016/S0091-679X(08)61905-6 [DOI] [PubMed] [Google Scholar]
- Tan J. L. and Zon L. I. (2011). Chemical screening in zebrafish for novel biological and therapeutic discovery. Methods Cell Biol. 105, 493-516. 10.1016/B978-0-12-381320-6.00021-7 [DOI] [PubMed] [Google Scholar]
- Taylor W. R. and Vandyke G. C. (1985). Revised procedures for staining and clearing small fishes and other vertebrates for bone and cartilage study. Cybium 9, 107-120. [Google Scholar]
- Thisse B. and Thisse C. (2014). In situ hybridization on whole-mount zebrafish embryos and young larvae. Methods Mol. Biol. 1211, 53-67. 10.1007/978-1-4939-1459-3_5 [DOI] [PubMed] [Google Scholar]
- Thisse C. and Thisse B. (2008). High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59-69. 10.1038/nprot.2007.514 [DOI] [PubMed] [Google Scholar]
- Topczewska J. M., Shoela R. A., Tomaszewski J. P., Mirmira R. B. and Gosain A. K. (2016). The morphogenesis of cranial sutures in zebrafish. PLoS ONE 11, e0165775 10.1371/journal.pone.0165775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turley S. D. (2004). Cholesterol metabolism and therapeutic targets: rationale for targeting multiple metabolic pathways. Clin. Cardiol. 27, III16-III21. 10.1002/clc.4960271506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlep Z., Lorbek G., Perse M., Jeruc J., Juvan P., Matz-Soja M., Gebhardt R., Bjorkhem I., Hall J. A., Bonneau R. et al. (2017). Disrupting Hepatocyte Cyp51 from Cholesterol Synthesis Leads to Progressive Liver Injury in the Developing Mouse and Decreases RORC Signalling. Sci. Rep. 7, 40775 10.1038/srep40775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter J. C., Adams M. D., Myers E. W., Li P. W., Mural R. J., Sutton G. G., Smith H. O., Yandell M., Evans C. A., Holt R. A. et al. (2001). The sequence of the human genome. Science 291, 1304-1351. 10.1126/science.1058040 [DOI] [PubMed] [Google Scholar]
- Vortkamp A., Lee K., Lanske B., Segre G. V., Kronenberg H. M. and Tabin C. J. (1996). Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science 273, 613-622. 10.1126/science.273.5275.613 [DOI] [PubMed] [Google Scholar]
- Wada N., Javidan Y., Nelson S., Carney T. J., Kelsh R. N. and Schilling T. F. (2005). Hedgehog signaling is required for cranial neural crest morphogenesis and chondrogenesis at the midline in the zebrafish skull. Development 132, 3977-3988. 10.1242/dev.01943 [DOI] [PubMed] [Google Scholar]
- Wassif C. A., Maslen C., Kachilele-Linjewile S., Lin D., Linck L. M., Connor W. E., Steiner R. D. and Porter F. D. (1998). Mutations in the human sterol delta7-reductase gene at 11q12-13 cause Smith-Lemli-Opitz syndrome. Am. J. Hum. Genet. 63, 55–62. 10.1086/301936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts S. A., Powell M. and D'Abramo L. R. (2012). Fundamental approaches to the study of zebrafish nutrition. ILAR J. 53, 144-160. 10.1093/ilar.53.2.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek D., Pawlik B., Li Y., Akarsu N. A., Caliebe A., May K. J., Schweiger B., Vargas F. R., Balci S., Gillessen-Kaesbach G. et al. (2010). A specific mutation in the distant sonic hedgehog (SHH) cis-regulator (ZRS) causes Werner mesomelic syndrome (WMS) while complete ZRS duplications underlie Haas type polysyndactyly and preaxial polydactyly (PPD) with or without triphalangeal thumb. Hum. Mutat. 31, 81-89. 10.1002/humu.21142 [DOI] [PubMed] [Google Scholar]
- Wray G. A. (2007). The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 8, 206-216. 10.1038/nrg2063 [DOI] [PubMed] [Google Scholar]
- Yang C., McDonald J. G., Patel A., Zhang Y., Umetani M., Xu F., Westover E. J., Covey D. F., Mangelsdorf D. J., Cohen J. C. et al. (2006). Sterol intermediates from cholesterol biosynthetic pathway as liver X receptor ligands. J. Biol. Chem. 281, 27816-27826. 10.1074/jbc.M603781200 [DOI] [PubMed] [Google Scholar]
- Yang J., Andre P., Ye L. and Yang Y. Z. (2015). The Hedgehog signalling pathway in bone formation. Int. J. Oral. Sci. 7, 73-79. 10.1038/ijos.2015.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X. (2001). A freely diffusible form of Sonic hedgehog mediates long-range signalling. Nature 411, 716-720. 10.1038/35079648 [DOI] [PubMed] [Google Scholar]
- Zeng X., Goetz J. A., Suber L. M., Scott W. J. Jr., Schreiner C. M. and Robbins D. J. (2001). A freely diffusible form of Sonic hedgehog mediates long-range signalling. Nature 411, 716-720. 10.1038/35079648 [DOI] [PubMed] [Google Scholar]
- Zhao Q., Eberspaecher H., Lefebvre V. and De Crombrugghe B. (1997). Parallel expression of Sox9 and Col2a1 in cells undergoing chondrogenesis. Dev. Dyn. 209, 377-386. [DOI] [PubMed] [Google Scholar]
- Zhao L., Chen X.-J., Zhu J., Xi Y.-B., Yang X., Hu L.-D., Ouyang H., Patel S. H., Jin X., Lin D. et al. (2015). Lanosterol reverses protein aggregation in cataracts. Nature 523, 607-611. 10.1038/nature14650 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.