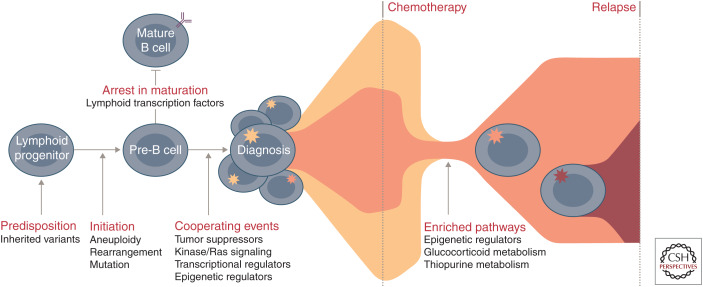
Figure 4.
Commonly altered pathways and stepwise progression of B-progenitor acute lymphoblastic leukemia (B-ALL). Common genetic polymorphisms (IKZF1, CDKN2A/B, PIP4K2A, GATA3, ARID5B, CEBPE, and ERG) and deleterious nonsilent inherited variants (PAX5, ETV6, IKZF1, TP53, and ERG) increase the risk of ALL susceptibility. Driving or founding lesions of ALL define genomic subtypes: aneuploidy and other chromosomal abnormalities (hyperdiploid, low-hypodiploid, near-haploid, iAMP21), rearrangements deregulating transcription factors (ETV6-RUNX1, ETV6-RUNX1-like, KMT2A, TCF3-PBX1, DUX4, ZNF384, MEF2D, NUTM1, TCF3-HLF, PAX5, BCL2/MYC) or kinase genes (Ph-like, BCR-ABL1), and specific mutations in lymphoid transcription factors (PAX5 P80R, IKZF1, N195Y). Deletion and loss of lymphoid transcription factors (e.g., IKZF1, PAX5, EBF1) coupled with the alteration of tumor suppressors and cell cycle regulators (CDKN2A/B, TP53), kinase signaling pathway genes (e.g., NRAS, KRAS, FLT3), other transcriptional regulators (e.g., ETV6, ERG), or epigenetic regulators (e.g., CREBBP, WHSC1, CTCF) result in the accumulation of immature lymphoid blasts and presentation at diagnosis. During treatment, the predominant diagnosis clone is commonly eradicated and relapse arises from a minor clone that already harbors and/or acquires additional genetic alterations that drive resistance. Pathways that are enriched at relapse include those involving epigenetic regulators (e.g., CREBBP, SETD2, KDM6A), the glucocorticoid response (e.g., CREBBP, NR3C1), and thiopurine metabolism (e.g., NT5C2, MSH6).