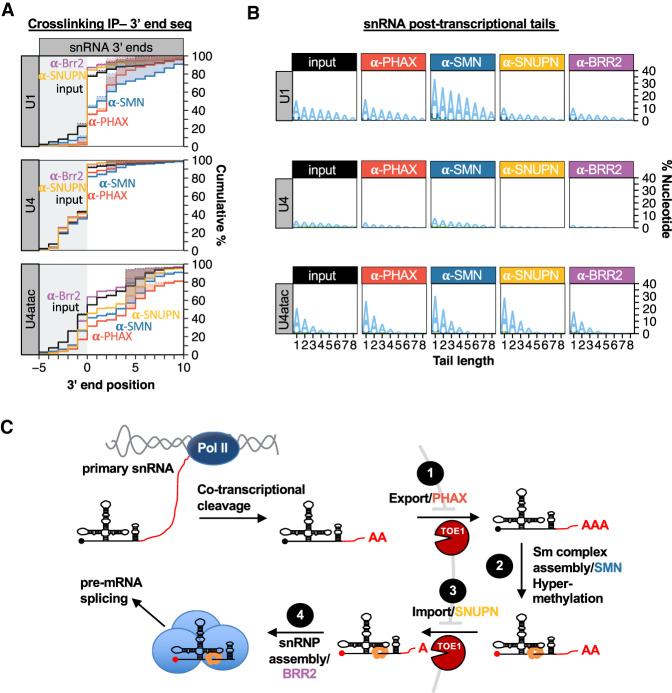
Figure 2.
snRNAs are tailed and trimmed at early and late steps of biogenesis. (A) Cumulative plots of 3′ end positions for snRNAs associated with snRNA biogenesis factors PHAX (red), SMN (blue), SNUPN (yellow), and BRR2 (purple), monitored by cross-linking and immunoprecipitation followed by snRNA 3′ end sequencing. Input samples are shown in black. For U4 snRNA, BRR2-associated 3′ ends are almost indistinguishable from those associated with SNUPN. Only reads terminating at or downstream from position −5 are represented. The average of three independent experiments is plotted for each snRNA. (B) Sequence logo plots representing the percent of biogenesis factor-associated snRNAs that contain posttranscriptionally added nucleotides in the presence or absence of TOE1, broken down by nucleotide composition. Averages of three independent experiments are plotted for each condition. (C) Schematic of snRNA biogenesis showing dynamic posttranscriptional tailing and trimming occurring at early and late steps. See also Supplemental Figure S2.