Figure 4.
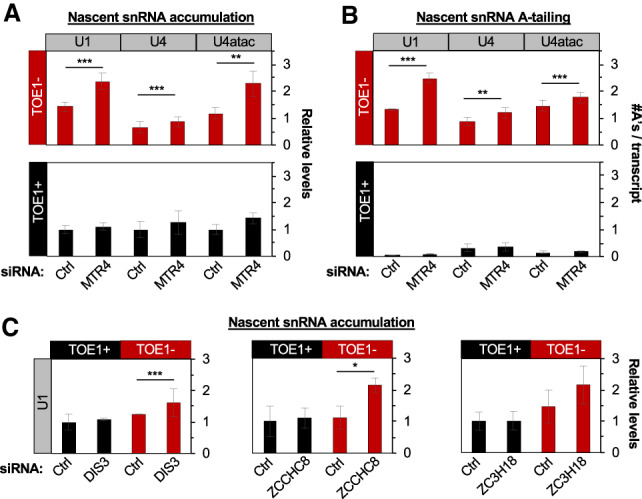
snRNAs become targets of the nuclear exosome in the absence of TOE1. (A) Relative levels of nascent snRNAs upon control (Ctrl) or MTR4 siRNA-mediated depletion in the absence (TOE1−) or presence (TOE1+) of TOE1. snRNA levels were measured by RT-qPCR and normalized to average snRNA levels in corresponding Ctrl siRNA/TOE1+ conditions (set to 1) with average levels of 7SK and mitochondrial 12S control RNAs serving as internal RT-qPCR normalization controls. (B) Average number of adenosines per snRNA transcript upon Ctrl or MTR4 siRNA-mediated depletion in the presence (TOE1+) or absence (TOE1−) of TOE1, monitored by 3′ end sequencing of nascent snRNAs. (C) Relative levels of U1 snRNA upon treatment with control siRNA (Ctrl) or siRNAs targeting nuclear exosome associated components, DIS3, ZCCHC8, and ZC3H18 compared with 7SK RNA and normalized to average snRNA levels in corresponding Ctrl siRNA/TOE1+ conditions. Error bars indicate SEM from at least three independent experiments for A and B and ZC3H18 in C and two independent experiments for DIS3 and ZCCHC8 in C. P-values (Student's two-tailed t-test): (*) P < 0.1; (**) P < 0.05; (***) P < 0.01. See also Supplemental Figure S4.
