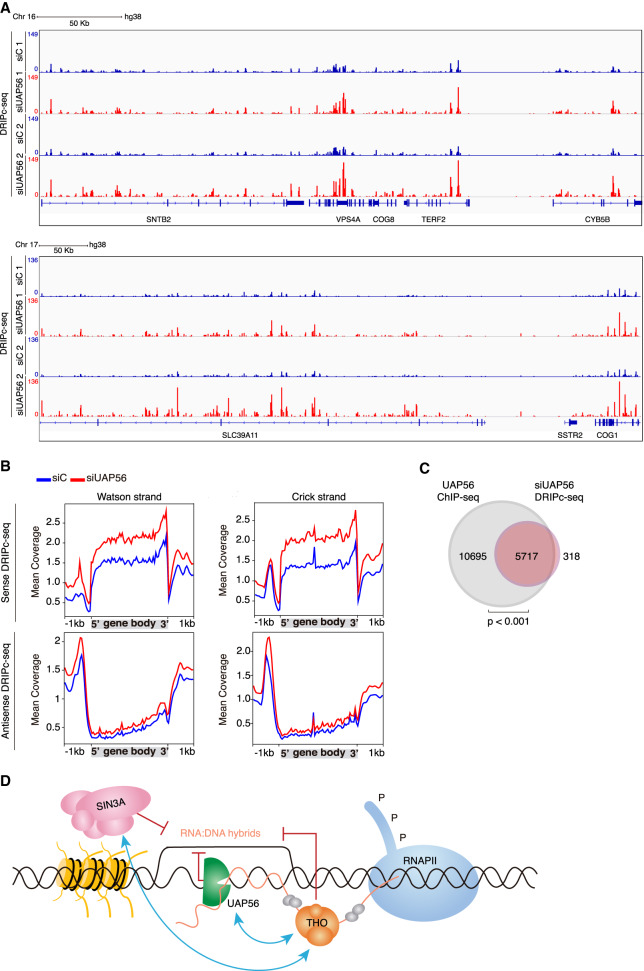
Figure 7.
Genome-wide accumulation of RNA–DNA hybrids in UAP56-depleted cells. (A) Representative screenshot of different genomic regions showing the DRIPc-seq signal profiles for siC (blue) and siUAP56 (red) (n = 2). (B) Distribution of antisense and sense DRIPc-seq signal (mean coverage) along a gene metaplot for siC control and siUAP56 cells over Watson (left) and Crick strand (right). (C) Venn diagram showing the overlap between R-loop gain genes (red) and genes to which UAP56 binds in normal K562 cells detected by ChIP-seq (gray). P < 0.001, hypergeometric test. (D) Model to explain the role of UAP56 in R-loop resolution during transcription. In the model, THO contributes to the proper assembly of the mRNP and talks to the Sin3A histone deacetylase complex to presumably transiently close the chromatin. The RNA–RNA and RNA–DNA helicase UAP56/DDX39B, apart from its role in RNA processing, ensures that unscheduled hybrids formed behind the transcription machinery are removed cotranscriptionally, thus potentially releasing the intact nascent RNA molecule for further processing and export.