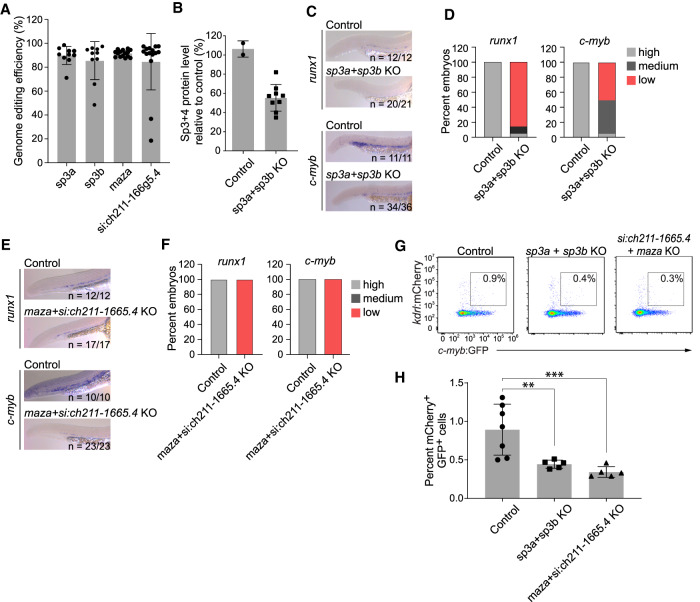
Figure 6.
Sp3 and Maz orthologs promote HSPC formation in zebrafish embryos. (A) Genome editing efficiency of each sgRNA determined using the tracking of indels by decomposition assay (TIDE). Data represent mean ± SD. n = 10 for sp3a; n = 10 for sp3b; n = 15 for maza; n = 15, for si:ch211-166g5.4. (B) Relative protein level of SP3 and SP4 in the knockout (KO) embryos. Because the only zebrafish antibody available is against both SP3 and SP4, the level of reduction of SP3 protein shown here is an underestimate. Data represent mean ± SD, n = 2 for-wild type embryos; n = 9 for sp3 knockout embryos. (C) Representative whole-mount in situ hybridization (WISH) images for runx1 and c-myb in the AGM region of 27 hpf and 30 hpf embryos, respectively. Embryos were injected with CRISPR guide RNAs targeting the Sp3 orthologs sp3a and sp3b. The numbers of embryos with reduced runx1 or c-myb expression relative to the number examined are indicated. (D) Bar graph depicting percentage of embryos with high, medium, and low runx1 and c-myb expression following injection of sgRNAs targeting Sp3 orthologs, and controls (summary of results in C). (E) Representative WISH images for runx1 (27 hpf) and c-myb (30 hpf) in the AGM region of control and KO embryos injected with CRISPR guide RNAs targeting both Maz orthologs, maza and si:ch211-166g5.4. (F) Percentage of embryos with high, medium, and low reporter gene expression after injection of sgRNAs targeting the Maz orthologs. (G) Representative flow cytometry showing gating strategy for mCherry+ GFP+ HSPCs from embryos transgenic for kdrl:mCherry and c-myb:GFP reporter genes following KO of Sp3 or Maz orthologs. (H) Frequency of mCherry+ GFP+ HSPCs in 30 hpf wild-type, Sp3, and Maz knockout embryos. n = 7 for wild-type embryos; n = 5 for Sp3 embryos; n = 5, for Maz knockout embryos; number of experiments = 2. (***) P ≤ 0.01; (**) P ≤ 0.05.