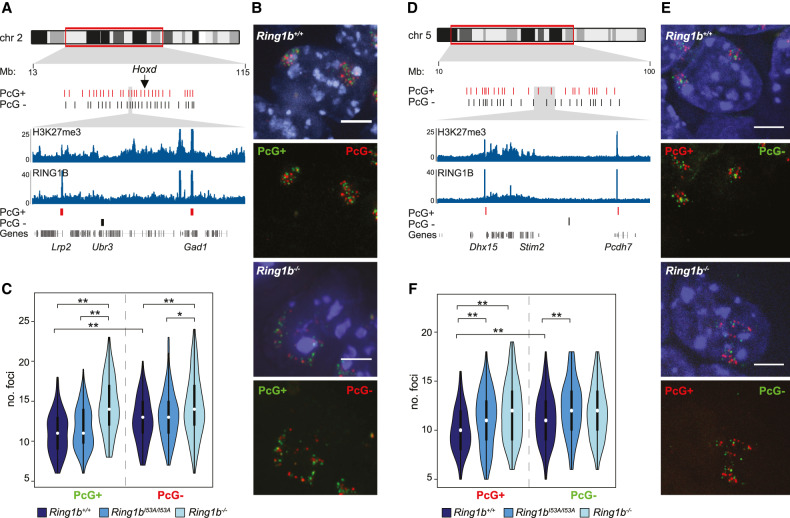
Figure 2.
Loss of PRC1 reduces nuclear clustering at sites with and without RING1B. (A) Ideogram of chromosome 2 indicating the location of the oligonucleotide probes used in B and C and zoomed in browser tracks of RING1B and H3K27me3 ChIP-seq from wild-type mESCs (Illingworth et al. 2015). Polycomb-positive (PcG+) and polycomb-negative (PcG−) are represented as red and black bars, respectively. Genomic locations are for the mm9 genome assembly. (B) Representative 3D FISH images of Ring1b+/+ and Ring1b−/− mESCs hybridized with the chromosome 2 oligonucleotide PcG+ (green; 6FAM) and PcG− (red; ATT0594) probes. Scale bar = 5 μm. (C) Violin plots depicting the number of discrete foci in Ring1b+/+, Ring1bI53A/I53A, and Ring1b−/− mESCs detected by the PcG+ and PcG− FISH probe pools. (*) P ≤ 0.05; (**) P ≤ 0.01; Mann Whitney test. (D–F) As for A–C, but for a second set of oligonucleotide probes targeted to chromosome 5. (E) PcG+ (red; ATT0594) and PcG− (green; 6FAM).