Fig 1. Generation of the PRISM Repurposing dataset.
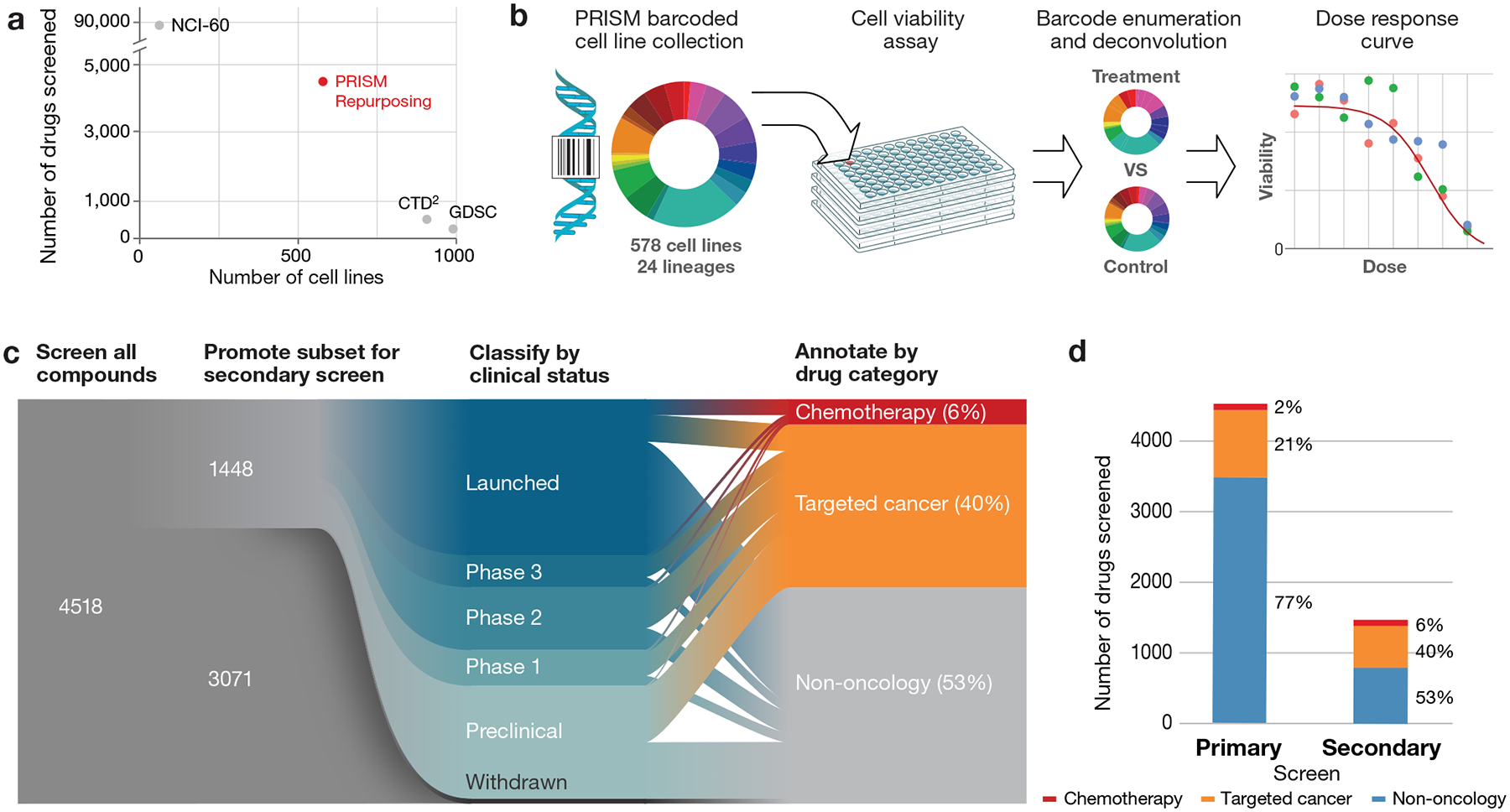
a, Dimensionality of publicly available pharmacogenomic drug screening experiments. The PRISM Repurposing dataset contains approximately ten-fold more compounds than CTD2 and approximately ten-fold more cell lines than NCI-60. b, PRISM method overview. Barcoded cell lines are pooled in groups of 25 and treated with chemical perturbagens. Pools are lysed 5days after perturbation and the relative abundance of mRNA barcodes is measured using Luminex MagPlex Microspheres to estimate cell viability. c, Repurposing screen workflow. A primary screen of 4,518 drugs was performed at 2.5 μM, followed by retesting of 1,448 active drugs at 8 doses. Compounds were annotated as chemotherapy drugs, targeted cancer drugs, or non-oncology drugs based on approved indications and prior clinical trial disease areas. d, Drug category representation in the primary and secondary screens. The secondary screen was enriched for chemotherapies and targeted cancer therapies.
