Figure 1. Conventionalization of germ-free mice induces the hepatic antioxidant and xenobiotic response.
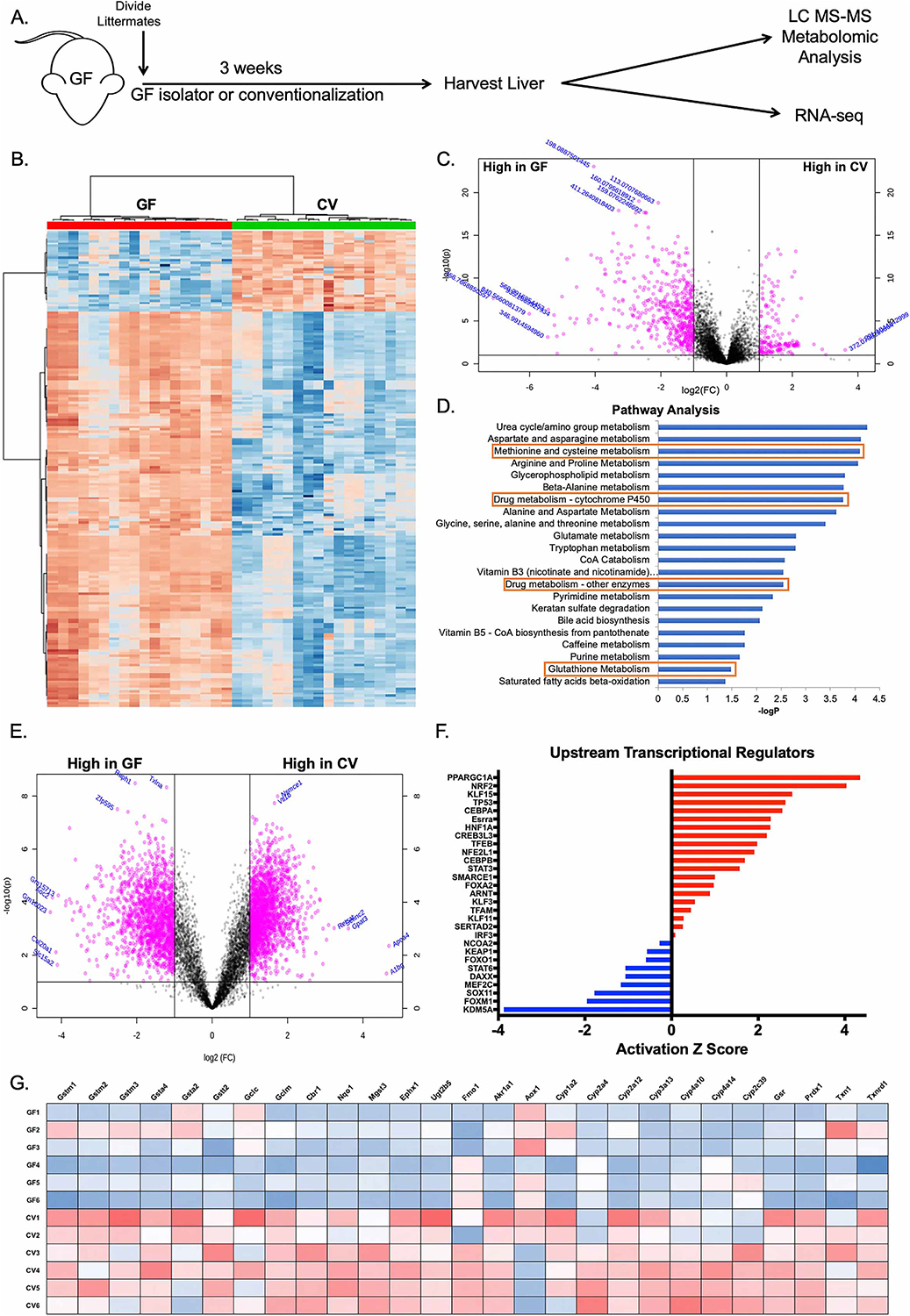
(A) Schematic of experimental design. Germ-free (GF) littermates were separated at weaning and either maintained GF or conventionalized (CV) with bedding from conventionally raised Specific pathogen free (SPF) mice for three weeks prior to sacrifice and harvest of liver tissue. (B) Hierarchical clustering of the top 200 altered features identified from ultra-high resolution LC-MS analysis of liver tissue from experiment described in (A). (C) Volcano plot of features identified in LC-MS analysis of liver tissue from experiment described in (A), where pink indicates p<0.05 as calculated by t-test. (D) Mummichog pathway analysis for the identification of pathways induced in CV relative to GF. Red boxes highlight pathways involved in antioxidant and xenobiotic metabolism. (E) Volcano plot of transcript enrichment detected by RNAseq analysis of liver tissue of GF and CV mice. Pink data points denote transcripts that were significantly enriched in GF or conventional mice to a confidence level of p<0.05 as determined by t-test. (F) Ingenuity Pathway Analysis of RNAseq identifies activated (red) and repressed (blue) transcriptional regulators in CV relative to GF mice. (G) Heatmap of genes under the transcriptional control of Nrf2 identified in RNAseq analysis. n=6 mice per condition (GF or CV), 3 male and 3 female.
