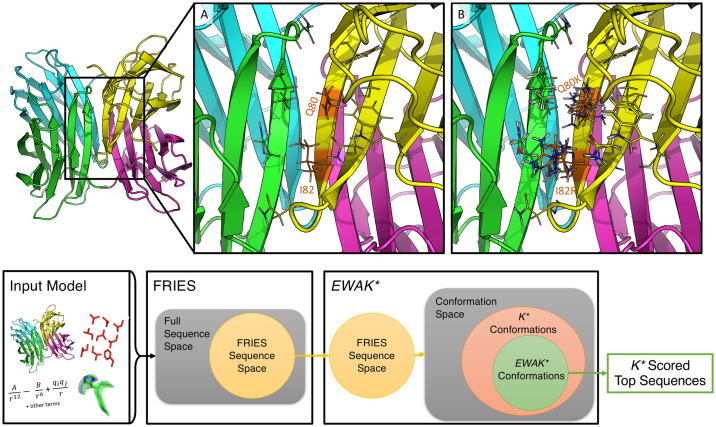
Fig 1. Design example using the structure of the LecB lectin Pseudomonas aeruginosa strain PA14 (PDB ID: 5A6Y [67]) and the osprey workflow for fries/EWAK*.
In the top panel, the full, 4 domain structure of lectin is shown on the left-hand side. (A) Zooming in on the region where domains A (green) and D (yellow) interact, showing the two mutable residues (Q80 and I82) along with the surrounding flexible shell of residues as lines. There were 11 flexible residues included in this design with Q80 and I82 allowed to mutate to all other amino acids except for proline. This design consisted of 8.102 × 1011 conformations and 441 sequences. fries limited this space to 5.704 × 1011 conformations and 206 sequences. fries/EWAK* in combination reduced the amount of time taken by about 75% compared to BBK*. fries alone was responsible for roughly 50% of this speed-up. (B) 10 low-energy conformations included in the thermodynamic ensemble of the design sequence with mutations Q80I and I82F. For this particular sequence, BBK* minimized 10,664 conformations while EWAK* minimized only 4,104 conformations. The bottom panel shows the general workflow for fries/EWAK*. The workflow begins with the input model (as described in the Section entitled “Computational materials and methods”), which defines the design space for the first algorithm, fries. fries proceeds to prune the sequence space as described in the Section entitled “Fast Removal of Inadequately Energied Sequences (fries)” and as illustrated in the Venn diagram with the unpruned space shown as a yellow disk. Next, the remaining fries sequence space defines the conformation space (which contains multiple sequences as well as conformations) searched with EWAK*. EWAK* limits the conformations included in each partition function as described in the Section entitled “Energy Window Approximation to K* (EWAK*).” EWAK* generally searches over only a subset of the conformations (green area) that previous K*-based algorithms like BBK* [32] search (orange area). EWAK* then returns the top sequences based on decreasing K* score.