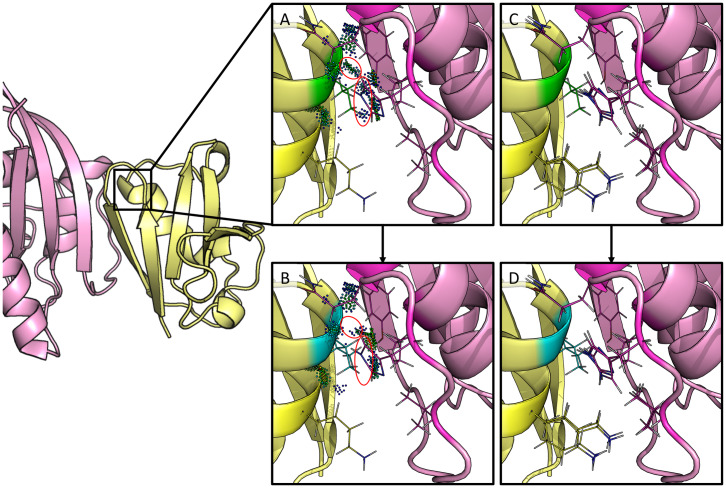
Fig 7. Redesign of c-Raf-RBD residue position 88 from valine to isoleucine.
The left-hand side shows c-Raf-RBD (yellow) in complex with KRas (pink). Panels (A-D) zoom in on one particular design at residue position 88 and are rotated 180°. Residue position 88 has a valine in the native, wild-type sequence (panels A & C) which was redesigned to an isoleucine (panels B & D). A mutation to isoleucine at this position was computationally predicted by EWAK* to decrease the binding of c-Raf-RBD to KRasGTP. This was experimentally validated in [60], where the authors incorrectly computationally predicted the effect of this particular mutation on the binding of c-Raf-RBD to KRasGTP. (A) The wild-type residue (valine) is shown in green with dots that indicate molecular interactions [71] with the surrounding residues (residues allowed to be flexible in the design are shown as lines). (B) The mutant residue (isoleucine) is shown in blue with dots that indicate molecular interactions [71] with the surrounding residues (residues allowed to be flexible in the design are shown as lines). Contacts made by the wild-type valine residue (circled dots in (A)) were lost upon mutation to isoleucine (circled space in (B)). (C & D) A set of 10 low-energy conformations that were included in the corresponding partition function calculation are shown for the wild-type (green) and the variant (blue).