Abstract
Identification of additional cancer-associated genes and secondary mutations driving the metastatic progression in pheochromocytoma and paraganglioma (PPGL) is important for subtyping, and may provide optimization of therapeutic regimens. We recently reported novel recurrent nonsynonymous mutations in the MYO5B gene in metastatic PPGL. Here, we explored the functional impact of these MYO5B mutations, and analyzed MYO5B expression in primary PPGL tumor cases in relation to mutation status. Immunohistochemistry and mRNA expression analysis in 30 PPGL tumors revealed an increased MYO5B expression in metastatic compared to non-metastatic cases. In addition, subcellular localization of MYO5B protein was altered from cytoplasmic to membranous in some metastatic tumors, and the strongest and most abnormal expression pattern was observed in a paraganglioma harboring a somatic MYO5B:p.G1611S mutation. In addition to five previously discovered MYO5B mutations, the present study of 30 PPGL (8 previous and 22 new samples) also revealed two, and hence recurrent, mutations in the gene paralog MYO5A. The three MYO5B missense mutations with the highest prediction scores (p.L587P, p.G1611S and p.R1641C) were selected and functionally validated using site directed mutagenesis and stable transfection into human neuroblastoma cells (SK-N-AS) and embryonic kidney cells (HEK293). In vitro analysis showed a significant increased proliferation rate in all three MYO5B mutated clones. The two somatically derived mutations, p.L587P and p.G1611S, were also found to increase the migration rate. Expression analysis of MYO5B mutants compared to wild type clones, demonstrated a significant enrichment of genes involved in migration, proliferation, cell adhesion, glucose metabolism, and cellular homeostasis. Our study validates the functional role of novel MYO5B mutations in proliferation and migration, and suggest the MYO5-pathway to be involved in the malignant progression in some PPGL tumors.
Author summary
Up to 25% of pheochromocytoma/paraganglioma (PPGL) cases develop metastatic disease with poor outcome and few treatment options. The disease mechanism is not fully understood, and to date there are no reliable markers to predict malignancy. We have recently discovered novel missense mutations in the non-conventional myosin 5 gene (MYO5B), an endosomal transport protein, which we now show enhances progression and migration in PPGLs. MYO5B mutations were preferentially found in patients with metastatic disease and SDH deficiency (germline SDHB-mutations). Abolished SDH activity result in a metabolic switch to aerobic glycolysis requiring increased glucose consumption. Since the MYO5B mutations were found to drive progression through downstream up-regulation of glucose metabolism genes, e.g. glucagon, we hypothesize that these mutations may fuel the pseudohypoxic state by altering glucose uptake in cancer cells. Our result is the first to link the myosin 5 genes to PPGL tumorigenesis. Further, it shows that the tumor progression route in PPGL is complex, with contribution from several genetic factors. An increasing number of studies show dysregulation and importance of the MYO5-proteins in cancer, but little is still known about the precise role and mechanism of mutations, hence more research in this area is needed.
Introduction
Pheochromocytomas (PCCs) and paragangliomas (PGLs), commonly denoted PPGLs, are rare neural tumors derived from chromaffin cells of the adrenal medulla and extra-adrenal paraganglia. PCCs and sympathetic PGLs secrete excess catecholamines with associated cardiovascular morbidity and mortality, and early diagnosis of these neoplasia is therefore vital. A great majority of PPGLs are sporadic and non-metastatic tumors and cured by surgery; however, up to 25% of cases develop metastatic disease (i.e. presence of PPGL in non-chromaffin organs) with poor outcome and few treatment options [1–4]. The survival rate at 5 years for patients with metastatic PPGLs is often less than 50% [5], and markers of metastatic disease are limited. PPGL have the highest degree of heritability in human neoplasms (around 30%), and over one-third of PPGLs are associated with inherited cancer susceptibility syndromes such as multiple endocrine neoplasia type 2 (MEN2), von Hippel–Lindau disease (VHL), neurofibromatosis type 1 (NF1), and hereditary paraganglioma (PGL1-PGL5). Inherited mutations have been identified in more than 14 genes, most commonly in VHL, SDHB, SDHD, NF1, and RET [6]. The most frequently mutated genes in PPGL belong to a wide range of functional classes, including kinase receptor and signaling (RET, NF1, HRAS, and MAX); energy metabolism (SDHA, SDHB, SDHC, SDHD, SDHAF2, FH); cellular response to hypoxia (VHL, and EPAS1 (also known as HIF2A)); endosomal signaling (TMEM127), and chromatin remodeling (ATRX). Germline and somatic mutations in these 14 susceptibility genes account for more than 60% of PPGL cases [7,8]. Several additional genes have been recently reported [4,9–15], and the identification of more tumor-driving genes and mutations in PPGL could facilitate diagnosis, treatment decision, and may provide new therapeutic options for patients [16].
Through large-scale sequencing of PPGLs, we recently reported recurrent mutations in the actin-dependent motor Myosin Vb gene; MYO5B [14] (Fig 1). Out of five novel nonsynonymous mutations identified, two somatically derived MYO5B variants (NP_001073936: p.L587P and p.G1611S) were found in metastatic sympathetic PGL cases from our data set [14]. Further screening of two public PPGL data sets [17,18] revealed three additional MYO5B mutations; one germline mutation in a metastatic PCC (p.R1641C), and two mutations in PCC tumor cases with metastatic potential (PASS score = 10) [19] or metastasized disease [18] (p.V1261G and p.D530E respectively). In addition, a novel somatic nonsynonymous mutation in isoform MYO5A (NP_000250.3: p.E926G) was discovered in a PCC tumor sample of our data set [14]. The non-conventional class V myosins, existing as three isoforms (Myo5A–C) in vertebrates, are motor proteins involved in intracellular cargo transport along actin-filaments. Class V myosins provide a molecular basis of a large number of essential cellular functions, such as cell motility, endocytosis, vesicle trafficking, and protein/RNA localization [20]. Myosin Vb (MYO5B) interacts with Rab-GTPases (Rab11, Rab8 and Rab11-FIP2), and plays an important role in vesicular transport and along the plasma membrane recycling pathway [21–23]. Loss-of-function mutations in MYO5B are common in microvillus inclusion disease (MVID) and cause disruption of cell polarity [24–27]. Emerging evidence show an important role of MYO5 proteins in several cancer types, where they often have an altered expression pattern [20,28]. For example, MYO5A expression is increased in a number of highly metastatic cancer cell lines and metastatic colorectal cancer tissues [29], and epigenetic downregulation of MYO5B has been reported to promote proliferation, invasion and migration in gastric cancer [30,31]. Additionally, MYO5B mutations and methylation-independent loss of MYO5B expression that matched disease progression was recently reported in colorectal cancer [32]. Mutations identified in PPGL cases are located in three domains of the MYO5B protein; the ATP dependent actin binding motor domain (p.D530E and p.L587P), coiled-coil rod domain mediating dimerization of motor protein and RAB8 binding (p.V1261G), and at the C-terminal globular tail domain that mediates cargo interactions and RAB11 binding (p.G1611S and p.R1641C) [33,34]. By functional prediction software, all five missense mutations were predicted to have an impact on the protein function, and none of the PPGL mutations were overlapping with previous reported MYO5B mutation spectra in colorectal cancer or MVID [14] (Fig 1). In this study we investigated the pathogenicity of the three most strongly predicted MYO5B mutations by functional in vitro studies. We also studied the protein and mRNA expression levels and subcellular location of MYO5B in primary PPGL tumors in relation to mutation status.
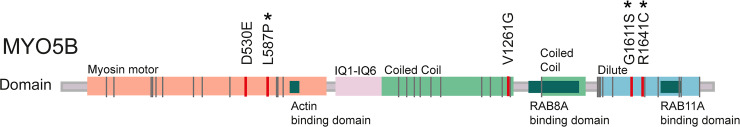
Fig 1. Localizations of mutations in MY05B.
Schematic presentation of the MYO5B gene, located on chromosome 18, containing 40 exons encoding the MYO5B protein (NP_001073936, 1848 amino acids). MYO5B constitutes of a myosin head domain and a globular tail (dilute) domain. Protein domains and motifs are presented according to Qiu et al [76], and the RAB8A- and RAB11A-binding sites according to Roland et al [23]. Localization of the five missense mutations in MYO5B identified in Wilzén et al [14] are marked in red, and the three selected mutations (p.L587P, p.G1611S, and p.R1641C) analyzed in the current study are marked with a star. Previously identified MYO5B mutations in colon cancer are according to Letellier et al [32] and marked in grey. IQ = IQ motif, BD = binding domain.
Results
Mutation status and MYO5B expression in primary PPGL tumors
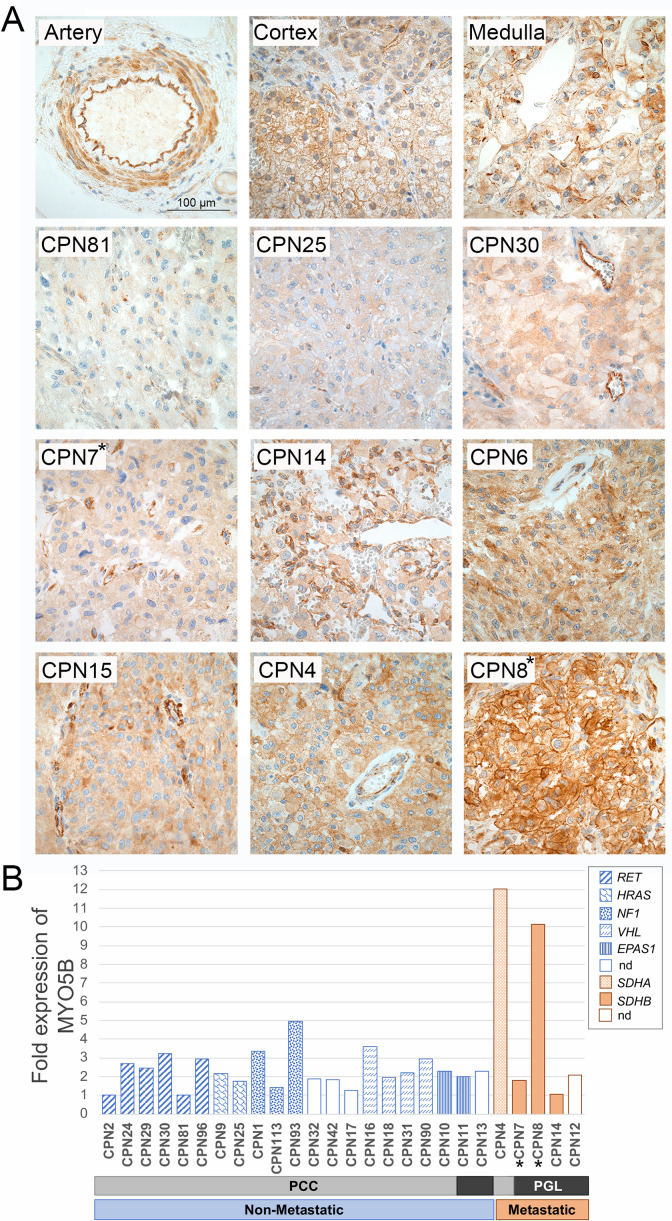
MYO5B expression levels were assessed in primary PPGL tumors by immunohistochemistry (IHC; 23 cases) and mRNA expression microarray (26 cases). (Table 1). IHC of three normal adrenal glands displayed a strong membranous MYO5B staining in cortex, and strong cytoplasmic staining of endothelial cells, while adrenal medulla displayed a weak cytoplasmic MYO5B protein expression (Fig 2A). The staining intensity and % of labeled tumor cells was scored from 0–9 according to Klein et al. [35]. Weak or negative cytoplasmic MYO5B expression was observed in 6/14 non-metastatic tumors (score <4), while 6/14 showed intermediate expression (score 4–6) and 2/14 displayed a strong expression (score>6; Table 1; Fig 2A). In contrast, metastatic tumors displayed a significantly stronger expression; 3/9 cases with intermediate expression and 6/9 displaying strong MYO5B expression compared to non-metastatic tumors (p = 0.007 by Mann-Whitney, 2-sided). Furthermore, a deviant membranous location of the MYO5B protein was found in three metastatic cases (CPN3, CPN4, and CPN8). The strongest and most aberrant MYO5B expression pattern was observed in case CPN8 harboring the p.G1611S MYO5B mutation (Fig 2A). However, case CPN7, harboring the p.L587P mutation, demonstrated an intermediate cytoplasmic MYO5B staining similar to the majority of PPGL cases (Fig 2A). Tyrosine hydroxylase (TH) staining of adjacent tumor sections is provided in S1 Fig. The mRNA expression analysis of 26 cases (20 non-metastatic, 5 metastatic, and 1 multifocal tumor; Table 1) confirmed a higher expression in metastatic compared to non-metastatic tumors (2-fold, p = 0.260, t-test, 2-sided). This difference was mainly due to two metastatic cases (CPN4 and CPN8) showing a 10-fold higher MYO5B expression than the non-metastatic cases, and interestingly, these were the same two cases showing an aberrant membranous staining by IHC (Fig 2B). Unfortunately, the mRNA expression of the CPN3 case could not be elucidated due to no fresh frozen tissue available.
Table 1. Patient data, mutation status, and MYO5B expression.
| Case ID | Sex | Diagnosis | Hereditary/ Sporadic | Syndrome | Metastatic/ Non-metastatic | Expr sub-type | Muta-tion status | MYO5B IHC | MYO5B mRNA Expr fold | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Expr Score | Localiza-tion | Expr Pattern |
|||||||||
| CPN2 | f | PCC | Hereditary | MEN2 | Non-M | 2 | RET | 6 | Cytoplasm | Granular | 1,00 |
| CPN24 | f | PCC | nd | NA | Non-M | 2 | RET | 6 | Cytoplasm | Granular | 2,68 |
| CPN29 | f | PCC | nd | NA | Non-M | 2 | RET | 3 | Cytoplasm | Even | 2,47 |
| CPN30 | m | PCC | nd | NA | Non-M | 2 | RET | 6 | Cytoplasm | Granular | 3,24 |
| CPN81 | f | PCC | Syndromic | MEN2 | Non-M | 2 | RET | 3 | Cytoplasm | Granular | 1,03 |
| CPN96 | f | PCC | Syndromic | MEN2 | Non-M | 2 | RET | 3 | Cytoplasm | Granular | 2,93 |
| CPN9 | m | PCC | Sporadic | NA | Non-M | 2 | HRAS | 2 | Cytoplasm | Patchy | 2,19 |
| CPN25 | f | PCC | nd | NA | Non-M | 2 | HRAS | 6 | Cytoplasm | Even | 1,74 |
| CPN1 | m | PCC | Sporadic | NA | Non-M | 2 | NF1 | 6 | Cytoplasm | Granular | 3,36 |
| CPN113 | f | PCC | nd | NA | Non-M | 2 | NF1 | nd | nd | nd | 1,42 |
| CPN93 | m | PCC | Syndromic | NF1 | Non-M | 2 | ni | nd | nd | nd | 4,94 |
| CPN32 | f | PCC | nd | NA | Non-M | 2 | ni | 0 | Cytoplasm | Even | 1,88 |
| CPN42 | f | PCC | nd | NA | Non-M | 2 | ni | nd | nd | nd | 1,82 |
| CPN17 | m | PCC | nd | NA | MuPC | 2 | ni | 6 | Cytoplasm | Even | 1,27 |
| CPN16 | f | PCC | nd | NA | Non-M | 1 | VHL | 9 | Cytoplasm | Granular | 3,61 |
| CPN18 | m | PCC | Hereditary | VHL | Non-M | 1 | VHL | 9 | Cytoplasm | Even | 1,94 |
| CPN31 | m | PCC | nd | NA | Non-M | 1 | VHL | 3 | Cytoplasm (neg) | Even | 2,20 |
| CPN90 | m | PCC | nd | NA | Non-M | 1 | VHL | nd | nd | nd | 2,94 |
| CPN10 | f | PCC | Sporadic | NA | Non-M | 1 | EPAS1 | nd | nd | nd | 2,31 |
| CPN11 | f | PGL | Sporadic, HPT | NE prod | Non-M | 1 | EPAS1 | nd | nd | nd | 1,99 |
| CPN13 | f | PGL | Sporadic | NA | Non-M | 1 | ni | nd | nd | nd | 2,29 |
| CPN4 | m | PCC | nd | DA prod | M | 2 | SDHA | 9 | Membrane | Granular | 12,04 |
| CPN8 | m | PGL | Hereditary | NA | M | 1 | SDHB | 9 | Membrane | Even | 10,15 |
| CPN3 | f | PGL | Hereditary | NA | M | nd | SDHB | 6 | Membrane | Patchy | nd |
| CPN6 | m | PGL | Hereditary | NA | M | nd | SDHB | 9 | Cytoplasm | Even | nd |
| CPN7 | m | PGL | Hereditary | NA | M | 1 | SDHB | 6 | Cytoplasm | Even | 1,79 |
| CPN12 | m | PGL | Sporadic | NA | M | 1 | ni | 9 | Cytoplasm | Even | 2,09 |
| CPN14 | m | PGL | Hereditary | NF1 | M | 2 | SDHB | 9 | Cytoplasm | Patchy & Granular | 1,04 |
| CPN15 | f | PCC | nd | NA | M | nd | ni | 9 | Cytoplasm | Patchy | nd |
| CPN123 | m | PGL | Hereditary | NA | M | nd | SDHB | 6 | Cytoplasm | Even | nd |
Patient data of 30 primary tumors and expression data of MYO5B by Immunohistochemistry (IHC) and mRNA microarray analysis. m = male, f = female, PCC = pheochromocytoma, PGL = paraganglioma, Hereditary = Family history or confirmed germline mutation in one of 14 PPGL major susceptibility genes, Sporadic = confirmed somatic mutation in one of 14 PPGL major susceptibility genes, Syndromic = Associated syndromic features, no known heredity, HPT = Primary hyperparathyroidism. NF1 = Neurofibromatosis 1, MEN2 = multiple endocrine neoplasia type 2, VHL = Von Hippel Lindau syndrome, NE prod = overproduction of norepinephrine, DA prod = overproduction of dopamine by biochemical analysis of urine and/or plasma. M = Metastatic (presence of tumor metastases in non-chromaffin organs), Non = Non-metastatic (no presence of tumor metastases), MuPC = Multifocal pheochromocytoma. Expr subtype = expression subtype by unsupervised hierarchical clustering: 1 = pseudohypoxic, 2 = kinase signaling. MYO5B IHC Expr Score = MYO5B protein expression level in tumor cells based on semiquantitative scoring system by Klein et al.[35], ranging between 0–9 (0 = negative, <4 = weak, 4–6 = intermediate, >6 = strong staining). Location (membrane, cytoplasm, or cell nuclei) and staining pattern (even, granular, or patchy) is provided. MYO5B mRNA expression fold = microarray expression level presented as fold change relative to the case with the lowest expression (case CPN2). ni = not identified, nd = not determined.
Fig 2. Expression of MYO5B in primary tumor tissue.
Immunohistochemistry (A) using MYO5B antibody of normal adrenal gland (top row), 3 representative non-metastatic PCC tumors (second row) and 6 representative PGL metastatic tumors (two bottom rows). Two tumors harboring somatic MYO5B mutations were included; CPN7 (p.L587P) and CPN8 (p.G1611S). Pictures are taken at 40 x magnification using a Nikon ECLIPSE E1000M microscope and a ProgRes C7 camera, scale bar shown is 100μm. mRNA expression analysis (B) by microarray in 26 PPGL cases. The normalized relative expression values are presented as fold change compared to the non-metastatic case with lowest expression (CPN2), and each tumor sample is presented with its mutation status and diagnosis; PCC (light grey), PGL (dark grey), non-metastatic (blue), metastatic (orange). The two cases harboring MYO5B mutations are marked with a star. NA = not available.
Mutation analysis of 40 genes by exome sequencing, and complementary Multiplex Ligation-dependent Probe Amplification (MLPA) analysis of 6 genes, identified a causative PPGL mutation in 23 out of 30 cases; 6 RET-mutated, 3 NF1-mutated, 4 VHL-mutated, 2 HRAS-mutated, 6 SDHx-mutated (SDHA, -B, -C, and -D), and 2 EPAS1-mutated tumors (Table 1; S1 Table and S2 Table). In the 7 remaining cases no apparent explanatory mutations among the 14 major PPGL susceptibility genes could be found. Mutations in RET, NF1, VHL, and HRAS were solely found in patients with PCC tumors, while mutations in SDH-genes were mainly found in patients with PGL tumors or metastatic PCC (Table 1). The majority of metastatic cases (7/9) harbored SDHB-mutations, one case displayed a germline SDHA- promoter mutation, and in one metastatic case no apparent PPGL-associated mutation could be found. The MLPA-analysis of CPN12, 13, 32, 42 and 93 displayed a complete somatic SDHB deletion (exon 1–8) at allele frequencies between 0.2 and 0.5. This pattern could either be due to loss of the SDHB-region, or chromosome 1p-deletion which is a classical somatic event in PPGL. Also, in CPN93 presenting syndromic NF1, a deletion of the whole NF1 gene was detected, This is most probably a secondary inactivation event occurring after a primary NF1 mutation, whichever could not be detected in the current analysis. In sample CPN81, a RET mutation (p.C609Y) was found at an allele frequency (AF) of 0.98, indicating a loss of heterozygosity of the wild type allele in the tumor tissue. In addition to the established PPGL susceptibility genes, secondary mutations were found in the following 8 genes; ARNT, DNAH17, MYCN, MYO5A, MYO5B, MYO9B, SLC25A11, VCL (S2 Table). Somatic mutations in 8 cases (CPN1-9) have been previously reported [14], and in the current study including 22 additional cases the following new mutations were identified: ARNT:p.Q391K, MYO5A:p.P1194Q, MYO9B:p.H1726Y and SLC25A11:p.A200S. Hence, by this study we have also identified the MYO5A gene as recurrently mutated in PPGLs; MYO5A:p.E926G previously identified in case CPN1 (somatic, AF = 0.29, [14]) and MYO5A:p.P1194Q identified in case CPN29 (somatic, AF = 0.25; S2 Table). In addition, a third MYO5A missense mutation, NM_000259.3: c.5065G>A, p.V1689I was identified in case CPN15 (germline, AF = 0.51), but this variant was filtered out due to presence in normal population (AF = 0.23% in Genome Aggregation Database (gnomAD) ALL). All three MYO5A mutations were predicted to be damaging by at least 2 out of 3 functional prediction algorithms (i.e. Polyhen, SIFT, and/or MutationTaster).
Unsupervised hierarchical cluster analysis of 153 genes with highest variance over samples, subdivided the 26 PPGLs samples into the two classical expression subgroups; cluster 1 comprising the Pseudohypoxic tumors (VHL/EPAS1-, and SDHx-mutated), and cluster 2 comprising the kinase signaling tumors (RET-, NF1-, and HRAS-mutated; S2 Fig, S3 Table) [36]. The only exception was the CPN14 case harboring a SDHB-mutation but with clinical manifestation of NF1, which was assigned to cluster 2. In addition, the CPN4 case, harboring a SDHA promoter mutation, showed a unique expression profile which did not fit well into any of the major PPGL expression subgroups (S2 Fig). The SDHA promotor mutation (c.-7A>C, p.?; S2 Table) has been described once before [37], but its pathogenicity is still uncertain.
Functional studies of stably transfected cell lines
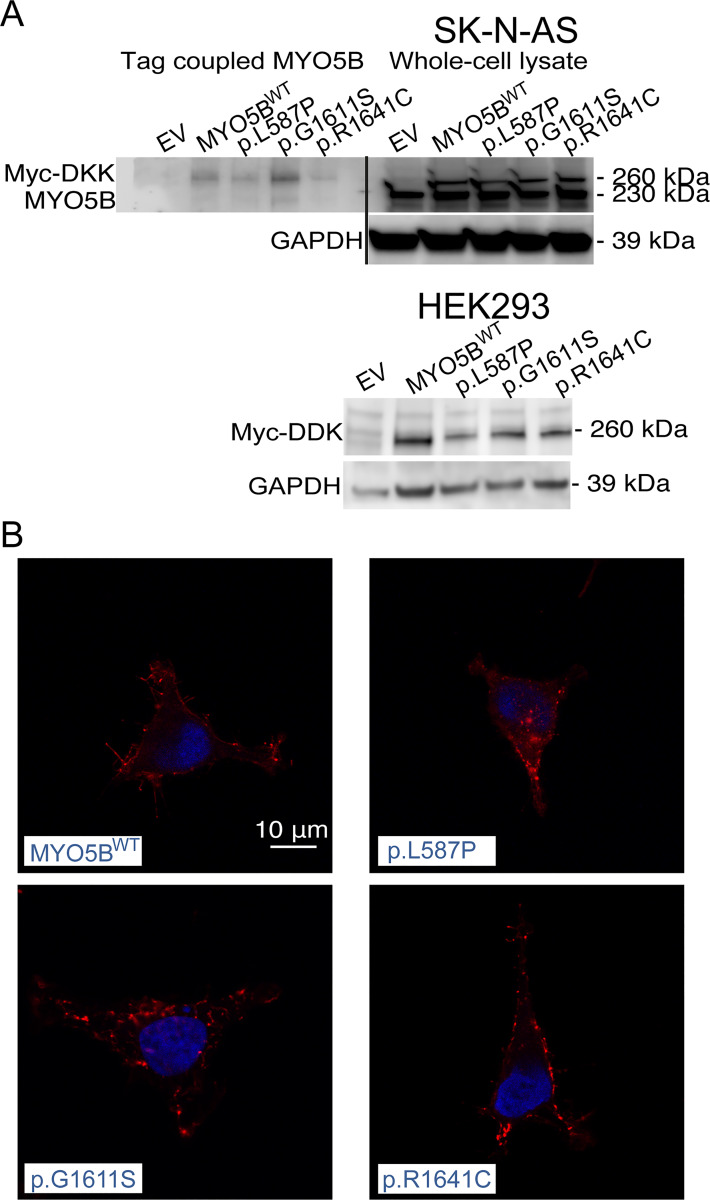
Three previously identified MYO5B missense mutations were selected based on theoretical prediction as damaging/deleterious/disease causing [14]; c.1760T>C, p.L597P; c.4831G>A, p.G1611S; c.4921C>T, p.R1641C (NM_001080467) (Fig 1). Stable clones of MYO5B mutants, MYO5B wildtype (WT) and empty vector constructs were generated in SK-N-AS and HEK293 cells. Western blot analysis of constructs showed a 230 kDa band representing endogenous expressed MYO5B protein, and 260 kDa band corresponding the additional size of the Myc-DKK-tag in transfected wildtype and mutated MYO5B proteins (Fig 3A). Co-immunoprecipitation of Myc-DKK-tagged proteins in SK-N-AS confirmed that 260 kDa band corresponded to the transfected MYO5B proteins (Fig 3A). By immunofluorescence the protein expression of mutated MYO5B (FLAG-tagged) showed a scattered cytoplasmic staining in small punctate spots, presumably localized to endosomal vesicles, in HEK293 cells (Fig 3B).
Fig 3. Expression and localization of MYO5B in cell lines.
Western blot in SK-N-AS cells (A) showing MYO5B expression in whole cell protein lysate and FLAG M2-antibody coupled lysate (n = 1, top panel). Western Blot using FLAG M2 antibody in HEK293 (n = 1, bottom panel). GAPDH used as loading control. Localization of Myc-DKK (B) in HEK293 (63X, LSM700) in pCMV6-MYO5B_L587P-Myc-DDK (p.L587P), pCMV6-MYO5B_G1611S-Myc-DDK (p.G1611S), and pCMV6-MYO5B_R1646C-Myc-DDK (p.R1641C). Myc-DKK is depicted in red and DAPI staining of nuclei in blue. Scale bar shown is 10μm.
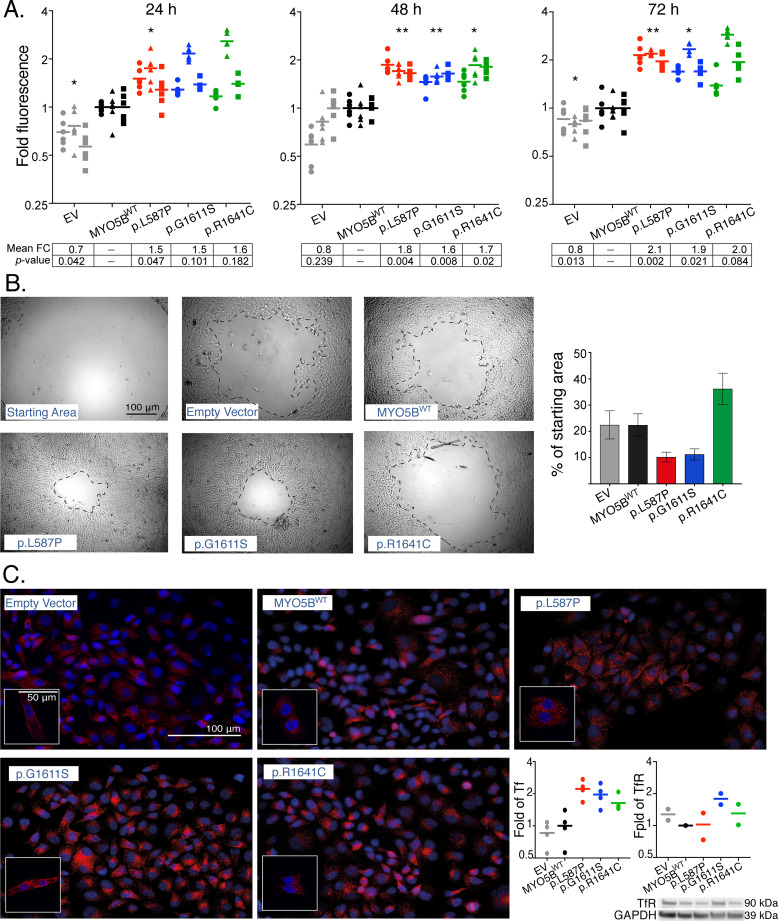
In order to assess the functional impact of the three MYO5B mutations in cultured, stably transfected cell lines, we investigated their ability to affect proliferation, migration and endosomal recycling. A significant increased proliferation (p<0.05) was found in all the three MYO5B mutations at 48h (p.L587P 1.8 fold; p.G1611S 1.6 fold; p.R1641C 1.7 fold), and the first and second mutations were also significant at 72h (p.L587P 2.1 fold; p.G1611S 1.9 fold) compared to MYO5BWT and empty vector clones (Fig 4A). The scratch-wound assay showed an increased migration rate of p.L587P and p.G1611S mutants with only 10% of the wound starting area remaining after 24 h, compared to MYO5BWT cells having 22% of the wound starting area remaining (Fig 4B). The p.R1641C mutation was not found to affect migration. The impact of mutations on endosomal recycling was explored by the transferrin trafficking assay. By visual inspection and fluorescence measurement of transferrin (Tf), the MYO5B mutations displayed a somewhat higher transferrin uptake (1.5–2 fold), most apparent in the cells harboring the p.L587P and p.G1611S mutations (Fig 4C). However, by relating the transferrin uptake to its receptor (TfR), the p.G1611S and p.R1641C mutants were found to express the transferrin receptor in almost the same proportion as transferrin, leaving p.L587P as the only mutation with slight impact on endosomal transport of transferrin (Fig 4C).
Fig 4. Proliferation, migration and endosomal recycling in MYO5B-mutants.
Proliferation with CyQuant NF assay in SK-N-AS clones (A) shown as fold change at 24h, 48h, and 72h compared to mean MYO5B wild type (WT). Graphs show three independent experiments, each run in sextuplicates. Grey = empty vector, black = MYO5BWT, red = p.L587P, blue = p.G1611S, green = p.R1641C. Mean fold change (FC) and significance (p-value) compared to MYO5BWT within each time point are presented (lower panel). * p<0.05, ** p<0.01, paired t-test. Migration (Oris) in HEK293 clones (B) shown as representable photos from light microscope (5X) of cell area remaining in MYO5B-mutants (p.L587P, p.G1611S, and p.R1641C), MYO5BWT, and empty vector at 24 h compared to the starting wound area. Scale bar shown is 100μm. Graph show percent wound area left after 24 h (mean±SEM, n = 2) in each construct. Transferrin uptake assay in SK-N-AS clones (C), with transferrin stained in red and nuclei stained in blue (DAPI). Pictures are taken with 40X Zeiss Axioscope 2 Plus fluorescence microscope, and small inserts show representative pictures taken with 63X, LSM700 confocal microscope. Scale bars shown are 50 μm and 100μm, respectively. Graphs plot fold change of corrected total cell fluorescence (CTCF) of transferrin (Tf) uptake (from four fields of view taken with 40X), and Western Blot analysis of the Transferrin receptor (TfR) expression (from two independent experiments) in mutants and empty vector compared to MYO5BWT (n = 2). GAPDH was included as a loading control.
Downstream expression analysis of MYO5B mutants
To elucidate the signaling pathways associated with MYO5B mutations in more detail, a transcriptome microarray analysis was performed in SK-N-AS constructs from three time points of proliferation (24h, 48h, and 72h) and from two replicated experiments. The global expression of each MYO5B mutant was compared to MYO5BWT and empty vector constructs for each time point, respectively. First, we checked and confirmed an up-regulated expression of the MYO5B gene in the clones harboring either mutant or wild type MYO5B cDNA vector constructs compared to clones with empty vector (S4 Table). Next, the top-ranked differentially expressed genes from all three MYO5B mutants were filtered out, and analyzed for enrichment of cellular processes using the Gene Ontology tool GOrilla (http://cbl-gorilla.cs.technion.ac.il) [38] (S5 Table). The gene ontology analyzes showed significant (p<0.001) enrichment of terms involving migration, i.e. “positive regulation of migration”, “cell motility”, “cellular component movement” and “protein kinase C activity” (S6 Table). Moreover, Gene Set Enrichment Analysis (GSEA) [39,40] was performed on each MYO5B mutant’s merged gene list (ranked after mean fold change) analyzing 236 Hallmark and KEGG curated gene sets from the Molecular Signature database (http://software.broadinstitute.org/gsea/msigdb/index.jsp). The most enriched gene sets in the three MYO5B mutants were the following four up-regulated: “Hallmark Myc targets v1”, “KEGG Base excision repair”, “KEGG Basal transcription factors”, “Hallmark E2F targets”, and four down-regulated: “Hallmark TNFA Signaling via NFKB”, “Hallmark Inflammatory response”, “KEGG ECM Receptor interaction”, “KEGG Arrhythmogenic right ventricular cardiomyopathy ARVC” (S7 Table).
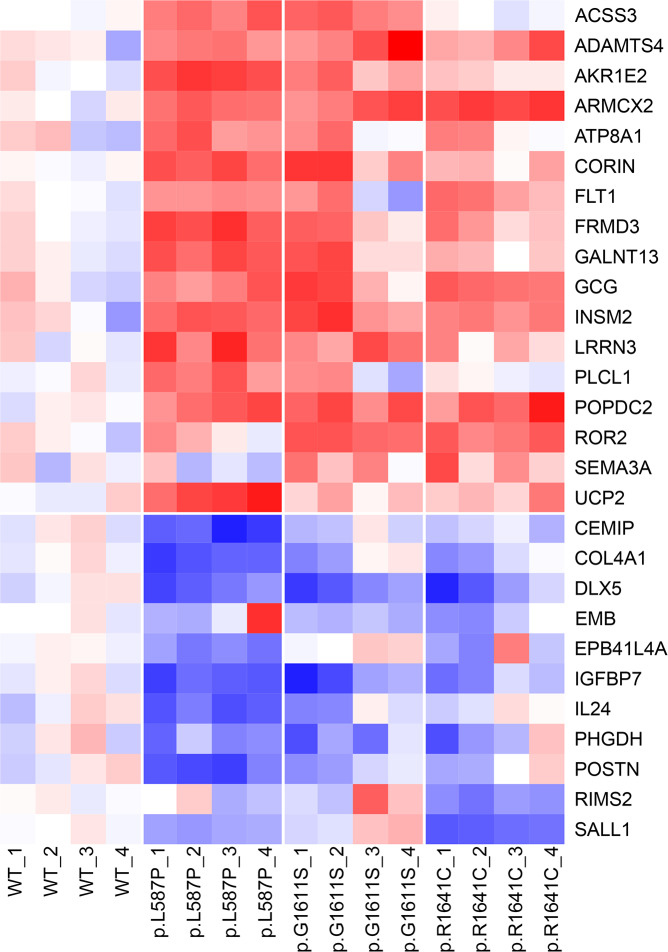
To verify the results, the 29 most differentially expressed genes (i.e. highest fold change) in all three mutants were selected from the top-ranked gene list; 17 up-regulated and 12 down-regulated genes compared to both MYO5BWT and empty vector (Table 2). These genes were run by TaqMan quantitative real-time PCR (qPCR) for both the same two samples used in the microarray analysis (passages p23 and p30 of SK-N-AS cells) and in two additional experiments (passages p21 and p28), all from proliferation time point 48h (Table 2). Overall, the expression pattern could be verified in all four experiments for each mutant, except for one gene (FILIP1L) which was below detection limit by qPCR (Fig 5; Table 2). The highest fold change (ΔΔCt >1.5 in at least 3/4 passages) for all three MYO5B mutants was found in three up-regulated genes: ARMCX2 (armadillo repeat containing X-linked 2), GCG (glucagon), INSM2 (insulin transcriptional repressor 2); and in four down-regulated genes: COL4A1 (Collagen Type IV Alpha 1 Chain), DLX5 (distal-less homeobox 5), IGFBP7 (insulin like growth factor binding protein 7), and POSTN (Periostin) (Fig 5; Table 2). Glucagon (GCG) and the insulin transcriptional repressor 2 (INSM2) were undoubtedly the two most up-regulated genes in all the three MYO5B mutants, with mean fold change of 7 and 14 respectively (Table 2).
Table 2. Differentially expressed genes in MYO5B-mutants.
| Gene symbol | Micro array |
qPCR verification | Gene name | Cellular function | |
|---|---|---|---|---|---|
| p23&p30 | p21&p28 | ||||
| GCG | 2,45 | 3,09 | 2,63 | glucagon | Glucose metabolism, homeostasis, and cell proliferation |
| INSM2 | 1,84 | 4,66 | 2,93 | INSM transcriptional repressor 2 | Glucose metabolism and homeostasis |
| GALNT13 | 1,51 | 1,62 | 1,28 | polypeptide N-acetylgalactosaminyltransferase 13 | Glycan biosynthesis and glycosylation |
| ARMCX2 | 1,33 | 1,88 | 1,92 | armadillo repeat containing, X-linked 2 | Mitochondria energy metabolism |
| FRMD3 | 1,07 | 1,19 | 1,09 | FERM domain containing 3 | Glucose metabolism and homeostasis |
| SEMA3A | 1,03 | 0,24 | 0,27 | semaphorin 3A | Neuronal and olfactory development |
| CORIN | 0,99 | 1,57 | 1,43 | corin, serine peptidase | Regulation of blood volume and pressure |
| FLT1 | 0,91 | 0,92 | 0,71 | fms related tyrosine kinase 1 | Angiogenesis and vasculogenesis, cell migration |
| FILIP1L | 0,88 | nd | nd | filamin A interacting protein 1 like | Focal adhesion, cytoskeleton, cell proliferation, and migration |
| ATP8A1 | 0,82 | 0,87 | 0,80 | ATPase phospholipid transporting 8A1 | Membrane transport, vesicle formation and trafficking, cardiac conduction, transport of glucose |
| ACSS3 | 0,81 | 1,12 | 1,14 | acyl-CoA synthetase short-chain family member 3 | Lipid synthesis and energy generation |
| UCP2 | 0,75 | 1,08 | 0,91 | uncoupling protein 2 | Mitochondria energy metabolism |
| AKR1E2 | 0,72 | 2,73 | 1,53 | aldo-keto reductase family 1 member E2 | Cell metabolism |
| PLCL1 | 0,62 | 0,47 | 0,49 | phospholipase C like 1 | Endocytosis process in neural cells |
| ROR2 | 0,61 | 1,65 | 1,29 | receptor tyrosine kinase like orphan receptor 2 | Cell proliferation, cell rigidity, bone formation |
| ADAMTS4 | 0,60 | 1,27 | 0,69 | ADAM metallopeptidase with thrombospondin type 1 motif 4 | Glycosylation, protein metabolism, cell migration |
| POPDC2 | 0,60 | 1,06 | 0,82 | popeye domain containing 2 | Heart rate dynamics |
| LRRN3 | 0,59 | 0,72 | 0,71 | leucine rich repeat neuronal 3 | Nervous system development |
| SALL1 | -0,68 | -1,09 | -1,50 | spalt like transcription factor 1 | Transcriptional regulation and organogenesis |
| PHGDH | -0,69 | -0,29 | -1,08 | phosphoglycerate dehydrogenase | Metabolism and electron transport activity |
| IL24 | -0,73 | -2,02 | -1,68 | interleukin 24 | Cell prolieration |
| EPB41L4A | -0,76 | -1,30 | -0,43 | erythrocyte membrane protein band 4.1 like 4A | Cytoskeleton and plasma membrane binding |
| EMB | -0,79 | -0,10 | -0,77 | embigin | Glucose metabolism and cell proliferation |
| RIMS2 | -0,90 | -0,43 | -0,19 | regulating synaptic membrane exocytosis 2 | Exocytosis and Rab3-binding |
| CEMIP | -1,35 | -1,63 | -1,65 | cell migration inducing hyaluronan binding protein | Glycosaminoglycan metabolism, cell migration, endocytosis |
| COL4A1 | -1,46 | -1,63 | -2,26 | collagen type IV alpha 1 chain | Focal adhesion, ECM, cell proliferation, migration |
| POSTN | -1,47 | -2,85 | -2,83 | periostin | Cell adhesion and migration, metastasis |
| IGFBP7 | -1,90 | -2,10 | -2,65 | insulin like growth factor binding protein 7 | Glucose metabolism and cell adhesion |
| DLX5 | -2,14 | -3,18 | -3,64 | distal-less homeobox 5 | Neural crest cell differentiation & cell proliferation |
Microarray mRNA expression levels and quantitative PCR (qPCR) verification results of the 29 most differentially expressed genes in SK-N-AS stably transfected MYO5B-mutants. Microarray expression data is presented as log2 fold change of MYO5B mutants (MUT 1 = p.L587P, MUT 2 = p.G1611S, MUT 3 = p.R1641C) compared to MYO5BWT, calculated from a mean of all three mutants from 2 passages of SK-N-AS (p23 and p30) at three time points (24h, 48h, and 72h). The qPCR data is presented as ddCt (log2 normalized expression to three endogenous control genes) from a mean of all three mutants compared to MYO5BWT at time point 48 hours of proliferation. Values from the same experiment as the micorarray (p23 and p30), as well as two additional experiments (p21 and p28), are presented.
Fig 5. Heatmap of differentially expressed genes in MYO5B mutants.
The heat map is based on qPCR data from 28 genes presented as dCt-values normalized gene-wise to a standard deviation equal to one, and the mean of the MYO5BWT samples subtracted. Data include three MYO5B mutants (p.L587P, p.G1611S, p.R1641C) and MYO5B wild type (WT) run as four independent experiments (passages 1 = p21, 2 = p23, 3 = p28, 4 = p30 of SK-N-AS), analyzed at time point 48h in each proliferation experiment. Red = up-regulated, blue = down-regulated.
Analyzing GCG or INSM2 in primary tumors harboring MYO5B mutations (i.e. CPN7 and 8) or in tumors with increased MYO5B expression (i.e. CPN4 and 8) showed no specific up-regulation of these genes. However, a heat map of the MYO5B mutation associated gene transcripts revealed a differential expression between the two major subtypes of PPGLs (S3A Fig). Also, the expression of MYO5-associated RABs (i.e. RAB8, RAB11, RAB11-FIP2, RAB25) were found to differentiate the two PPGL clusters (S3B Fig).
Discussion
Sequencing efforts and integrative genomic studies have started to uncover the genetic landscape of abnormalities that underlie PPGL tumors. However, functional verification work is needed to pinpoint the pathogenic disease-driving alterations in these tumors. We have previously published a study identifying novel recurrent MYO5B mutations in metastatic PPGL [14]. Here, we functionally verified the tumorigenic properties of three of these mutations by in-vitro studies in SK-N-AS and HEK293 cell lines. The human neuroblastoma cell line SK-N-AS was selected since it shares the same neural-crest origin, is wild type for all currently known PPGL susceptibility genes, and successfully used in several previous PPGL studies requiring a cell line of human origin [41–44]. Also, analyses in primary tumors revealed differential expression and altered sub-cellular localization of the MYO5B protein in metastatic PPGL, and mutation screening of additional PPGL cases identified recurrent mutations in the MYO5A paralog.
MYO5B is an actin-dependent intracellular vesicle transport myosin protein that has essential functions in the regulation of intracellular transport, membrane recycling, and cell movement. Mutations in MYO5B have been shown to disrupt cellular polarity in MVID [24,25]. Cell polarity is crucial for normal physiology and also plays a major role in development of tumor metastasis [45]. A link between the protein isoform MYO5A and cancer has already been established [29,46]. MYO5A show elevated expression in metastatic cancer cell lines derived from various tissue types, and is connected to tumor cell migration and metastasis in vitro and in vivo [29]. Emerging evidence also suggest that MYO5B proteins have an important role through differential expression in multiple cancer types [20,47]. Low protein levels of MYO5B has been shown to be associated with motility of gastric cells [30], and expression level and mutations in MYO5B has been reported as a powerful prognostic biomarker in colorectal cancer, which might help to stratifying patients for adjuvant therapy [30,32]. Here, we show a significantly stronger expression of MYO5B protein in metastatic tumors and much elevated mRNA expression levels (10-fold) in a subset of metastatic PPGL tumors. Moreover, an altered subcellular localization of the MYO5B protein to the membrane was observed in three metastatic cases, and the most prominent abnormal pattern was observed in the tumor case harboring the p.G1611S mutation. Although the exact mechanism of the altered protein localization is not known, we speculate that this could reflect a dysregulated exocytosis or endocytosis in these metastatic PPGLs. Nevertheless, the divergent pattern implies a role of MYO5B in the metastatic progression route.
The tumorigenic properties of the MYO5B mutations were demonstrated by a significantly increased proliferation rate for all three missense mutations (p.L587P, p.G1611S and p.R1641C). Furthermore, the p.L587P and p.G1611S mutations both showed increased migratory properties, and consistently the most significantly enriched GO-term among the differentially expressed genes in MYO5B-mutants was “positive regulation of migration”. With regard to the p.R1641C, it appears that this mutation had less effect on proliferation compared to the other two MYO5B mutations, and also did not increase migration. This is possibly due to the fact that p.R1641C was identified as a germline mutation in the primary case, while p.L587P and p.G1611S were originally discovered as somatic mutations [14, 17]. MYO5B is also implicated in regulating different pathways for endosomal recycling of proteins to the plasma membrane through interaction with Rab11, Rab8 and Rab11-FIP2 [21–23,48]. Although a moderately higher transferrin uptake in all three MYO5B mutants was observed, only p.L587P showed a slight effect on transferrin uptake after normalization to the transferrin receptor expression. Since the p.L587P is located in the actin binding myosin head, while p.G1611S and p.R1641C are located in the tail domain mediating Rab11 binding, the mechanisms may differ, which could also explain the different influence on transferrin receptor expression [23]. Taken together, the functional in vitro studies of MYO5B mutations suggests a gain of function or a dominant negative role of the mutants, enhancing mainly proliferation and migration.
The downstream transcriptome analysis of transfected SK-N-AS MYO5B-mutants showed a differential expression of genes involved in cell growth and proliferation, cell migration, cell adhesion, endosomal transport and glucose metabolism. Among the top-ranked down-regulated genes, DLX5 (distal-less homeobox 5), has previously been found among the genes with the highest number of hypermethylated CpG sites in primary metastatic neuroblastoma tumors [49]. Also, IGFBP7 (insulin-like growth factor binding protein 7), has been reported as epigenetically down-regulated and an independent prognostic factor in gastric cancer, leading to increased cell growth, invasion and migration [50]. Moreover, deletion of IGFBP7 was found to increase proliferation in hepatocellular carcinoma by a constitutively active IGF signaling [51]. Interestingly the third most downregulated gene, POSTN (Periostin), is a multifunctional glycoprotein that plays a role in the adhesion process, in the migration of many cells, and importantly, in the epithelial-mesenchymal transition of cancer cells. Periostin has been linked to renal cell carcinoma, where it promotes migration and invasion via the integrin/focal adhesion kinase/c-Jun N-terminal kinase pathway [52]. Periostin has a prognostic value in multiple solid cancers, and has been suggested as a potential therapeutic target in human solid cancer [53]. Most conspicuous was the 14-fold increase in expression of INSM2 (insulin transcriptional repressor 2) and the 7-fold increase in expression of GCG (glucagon) in all three MYO5B mutants. The pancreatic islets regulate glucose metabolism through secretion of islet hormones such as insulin and glucagon, and INSM2 is a direct target of Ngn3 and NeuroD1, two crucial transcriptional factors involved in human diabetes and pancreatic islet development [54]. Deletion of Insm2 in mice resulted in reduced insulin secretion and glucose intolerance [55]. INSM2 is also called Insulinoma-associated gene 6 (IA6); Insulinomas are rare neuroendocrine tumors of the pancreas that are usually sporadic, but may occur in association with multiple endocrine neoplasia type 1 (MEN1) syndrome [56]). Also, glucagon signaling is shown to significantly stimulate proliferation in colon cancer cell lines [57]. Metformin, one of the most commonly used insulin sensitizers, has been demonstrated to have anti-proliferative effects in several human malignancies [58,59]. Furthermore, metformin has recently been shown to suppress proliferation in rat pheochromocytoma cell line PC12 [60]. Thus, the increased proliferation rate seen in the MYO5B mutants might be driven through altered energy metabolisms.
Interestingly, the glucagon receptor (GCGR) was found among the 153 classifier genes with the highest variance between PPGL subtypes, and has previously been reported as discriminative between SDHB- deficient tumors (high expression) and VHL- deficient tumors (low expression) [61], which is in line with our data. Altered metabolism is a key feature of the pseudohypoxic PPGLs [62] and has been linked to the Warburg effect with increased aerobic glycolysis [63]. The alternative energy-generation pathway is somewhat less efficient, requiring a much larger cellular influx of glucose to maintain the energy needs in tumor cells, and the increased glucose consumption can be exploited for diagnostic purpose [64]. Provocative tests using glucagon has previously been performed in patients with undiscovered PPGLs, and these tests have been reported to lead to multi-organ failure or hypertensive emergency in some cases [65–67]. This, together with the low sensitivity of the glucagon test, and diverse expression of the glucagon receptor in different PCC syndromes has led to recommendations to not use this test in clinical practice [68]. In light of our results, the possible direct involvement of glucagon in increasing proliferation and in fueling the glucose-dependent oncogenic state in specific subtypes of PPGLs, especially SDH-deficient tumors [69], might add to this list of arguments.
Conclusion
An increasing number of studies show dysregulation of the MYO5-pathway in tumorigenesis and malignancy. In the present study we have functionally verified the tumorigenic role of three novel MYO5B mutations through their impact on proliferation and migration. The high expression and altered subcellular localization of MYO5B protein in a few malignant tumors speaks for an important role in progression of PPGL. Also, this study identified additional recurrent mutations in the MYO5A paralog, adding evidence of the MYO5-pathway's involvement in the tumorigenesis of PPGL. In conclusion, our study adds deeper insight into the complex genotype-phenotype correlation in PPGLs and also augments the emerging evidence of MYO5B's involvement in cancer.
Materials and methods
PPGL patients and primary tumor data
Fresh snap-frozen (SF) tumors (27 samples) and formalin-fixed paraffin-embedded (FFPE) tumor sections (3 samples) from 30 PPGL patients that underwent surgical resection between years 2000–2017 at the Department of Surgery, Sahlgrenska University Hospital, Gothenburg, Sweden, were included in the study (Table 1). Corresponding germline samples (blood or FFPE sections) were included for 15 of the patients. All tissue samples underwent routine pathological examination.
Ethics Statement
The study was approved by the ethical review board in Gothenburg (Dnr: 239–13, Approved May 2013). All patients gave their written consent prior to surgery.
Immunohistochemistry of primary PPGL
Paraffin embedded tumor sections and normal adrenal tissue (3–4 μm) were deparaffinized, rehydrated and antigen retrieved in DAKO PT Link using EnVision FLEX Target Retrieval Solution (Dako). Immunohistochemistry (IHC) staining of tumor sections with antibody against MYO5B (1:250, Sigma) was performed in Dako Autostainer Link using EnVision FLEX with EnVision FLEX+ (LINKER) according to the manufacturer’s instructions (DakoCytomation). The stained sections were photographed using a Nikon ECLIPSE E1000M microscope with 40x objective and a ProgRes C7 camera. The scoring of MYO5B staining was evaluated independently by a board certified surgical pathologist (ONW) and a clinical geneticist (FA) using an Olympus BX51 light microscope. The MYO5B protein expression in tumor cells and adrenal tissue was scored based on the intensity of staining and % of stained cells according to the combinative semiquantitative scoring system provided by Klein et al.[35]. The scoring system range between 0–9 obtained from multiplication of the two following point scales: % cells (0% = 0, <33% = 1, 33–67% = 2, >67% = 3) and intensity (negative = 0, weak = 1; mild = 2, strong = 3). Moreover, the location (membrane, cytoplasm, or cell nuclei) and staining pattern (even, granular, or patchy) was inspected. The intensity of tumor sections was scored as strong when it had the same intensity as cortex and artery in the normal adrenal tissue and staining in more than 2/3 of the tumor cells (score 3*3 = 9). Immunostaining of Tyrosine hydroxylase (clone 1B5 from Leica Biosystems) in adjacent tumor sections was performed according to standard procedures in clinical routine to confirm the diagnosis of PPGL, and slides were scanned using a LEICA SCN400 scanner (S1 Fig).
Mutation and expression analysis of primary PPGL
Genomic DNA from tumor tissue and blood was extracted according to Wilzén et al [14]. Mutation analysis of 40 target genes was performed by Illumina sequencing on 30 cases in either pair of tumor tissue (T) and normal tissue/blood (N) (15 cases) or as single tumor tissue (15 cases, S1 Table and S2 Table) [4,7,14,18,70–73]. Sequencing of cases CPN1-9 has previously been described [14]. The 22 new cases were sequenced by SureSelect v3/v5 (Agilent Technologies, CA) with paired-end (2*75-100bp) on a HiScanSQ (CPN10-15) or by SureSelect Clinical Research Exome v2 (CREv2; CPN16-CPN123), paired-end (2*100bp) on a NextSeq500 illumina sequencer. For details on variant calling and assessment see supplementary methods (S1 File). Only variants predicted to be damaging by functional prediction algorithms and/or previously reported in ClinVar (www.ncbi.nlm.nih.gov/clinvar/) or HGMD (portal.biobase-international.com) databases were included in the final mutation list (S2 Table). Sequence data has been deposited at the European Genome-phenome Archive (EGA), which is hosted by the EBI and the CRG, under accession number EGAS00001001601 (8 previous PPGL samples) and EGAS00001003991 (22 new PPGL samples). Further information about EGA can be found on https://ega-archive.org and "The European Genome-phenome Archive of human data consented for biomedical research"(http://www.nature.com/ng/journal/v47/n7/full/ng.3312.html). Nine cases with no apparent disease-associated pathogenic mutation by sequencing where further analyzed for exon/gene deletions or duplications by MLPA in 6 genes; SDHA, SDHB, SDHC, SDHD, NF1, and VHL. MLPA was performed on tumor DNA using the following four SALSA MLPA kits; P081-D1 & P082-C2 (NF1), P226-D1 (SDHx), and P016-C2 (VHL) (MRC-Holland www.mrcholland.com).
Expression analysis was performed on total-RNA from fresh tumor tissue by 44K Agilent Cy3/Cy5 2-color microarrays (see S1 File for details). Unsupervised hierarchical clustering of both samples and genes was performed using Omics Explorer 2.0 Beta from Qlucore (www.qlucore.se) using the Average linkage of Euclidian metric (each variable was normalized to mean = 0 and variance = 1). Using the filtering variance slider, transcripts with the lowest variance were filtered out until distinct subgroups appeared, resulting in a set of 153 genes (156 variables; S3 Table). Samples were divided into two cluster groups based on the dendrogram.
MYO5B vector construct design
The full coding sequence of MYO5B wild type (NM_001080467) and MYO5B sequences harboring the three point mutations c.1760T>C (p.L597P), c.4831G>A (p.G1611S) and c.4921C>T (p.R1641C) were synthesized and cloned into the pCMV6 vector, and tagged with Myc-DKK tag by Invitrogen GeneART (Thermo Fisher Scientific). Five vector constructs; pCMV6-Myc-DDK (empty vector), pCMV6-MYO5B-Myc-DDK (MYO5BWT), pCMV6-MYO5B_L587P-Myc-DDK (p.L587P), pCMV6-MYO5B_G1611S-Myc-DDK (p.G1611S), and pCMV6-MYO5B_R1646C-Myc-DDK (p.R1641C) were sub-cloned and verified by DNA Sanger sequencing (GeneArt Gene Synthesis, Invitrogen, ThemoFisher Scientific).
Cell culture and stable transfection
Human cancerous neuroblastoma cells (SK-N-AS, European Collection of Authenticated Cell Cultures, ECACC, SigmaAldrich) sharing the same embryologic neural crest origin as PPGL, and the non-cancerous embryonic kidney cells (HEK293, human embryonic kidney cell line, American Type Culture Collection, ATCC, USA), were cultured in high glucose DMEM (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (HEK293, Thermo Fisher Scientific) or 10% HyClone bovine growth serum (SK-N-AS, Thermo Fisher Scientific) in 37°C and 5% humidified CO2 using standard procedures. Prior to transfection, 4x 105 cells were seeded onto 6-well plates one day ahead. Transfection of pCMV6-Myc-DDK vector constructs into SK-N-AS and HEK293 cells was performed using 2.5 μg of each construct, 7.5 μl Lipofectamine 3000, and 5 μl P3000 reagent (Thermo Fisher Scientific) following the manufacturer’s protocol. Five constructs per cell line were made; MYO5BWT, p.L587P, p.G1611S, p.R1641C, and empty vector. When cells reached confluency, they were seeded onto 10 cm plates and selection medium (500 μg/ml Geneticin, G418, Thermo Fisher Scientific), was added to the cells. Cells were sub-cultured with selection medium until experiments were conducted.
Protein preparation and Western Blot
Transfected SK-N-AS and HEK293 were harvested and lysed using RIPA-buffer supplemented with phosphatase- and protease- inhibitors (Thermo Fisher Scientific). Protein lysates (30 μg) were resolved on 4–20% precast gels (Bio-Rad Laboratories) and transferred onto 0.45 μm PVDF membranes (Thermo Fisher Scientific). Western blot was performed using antibodies with ECL-detection (Supersignal West Maximum Fempto, Thermo Fisher Scientific) as follows: FLAG-DDK M2 mouse mAb (1:750, #F3165, Sigma Aldrich), GAPDH rabbit Ab (1:500, sc-25778, Santa Cruz Biotechnology), MYO5B Rabbit mAb (1:250, HPA040902, Atlas Antibodies), transferrin receptor (CD71) rabbit mAb (1:500, #13208, Cell Signaling Technology). Chemiluminescent signal from membranes were imaged using a LAS-400 imaging system (Fujifilm). The experiment was performed twice and quantified using Image Studio Lite v5.2.5 (https://www.licor.com/bio/image-studio-lite/). GAPDH was used as a loading control.
Co-Immunoprecipitation
Proteins from SK-N-AS were co-immunoprecipitated using the Dynabeads co-immunoprecipitation kit (Thermo Fisher Scientific). FLAG-DKK M2 mouse antibody (1:100, Sigma Aldrich) was coupled to Dynabeads M-270 Epoxy beads. Antibody coupled beads (1.5 mg) and 1 mg protein lysate was used in the co-immunoprecipitation in order to form protein complexes. Co-immunoprecipitated proteins from SK-N-AS were then subjected to western blot as described above using MYO5B antibody in order to confirm FLAG-DKK/MYO5B association.
Localization of FLAG-DKK
Transfected HEK293 cells were seeded at 2.5x 104 cells/well onto MilliCell EZ slides (Merck Millipore) and allowed to attach during 48 h. Cells were fixed using 4% paraformaldehyde (HistoLab) and washed with Dulbecco’s PBS (+Mg2+/+Ca2+, Gibco, Life Technologies). Chambers were pretreated with AB-buffer containing 1% BSA, 0.5% Triton-X diluted in PBS during 5 minutes and stained with antibodies as follows: FLAG-DDK M2 mouse mAb (1:500), and Alexa Fluor 555 anti-mouse IgG (1:1000, A21424, Life Technologies) Chamber slide walls were detached and slides were mounted with Prolong Gold (Invitrogen, Life Technologies). Localization of proteins were visualized (63X) using LSM700 confocal microscope (Carl Zeiss).
Cell proliferation
Transfected SK-N-AS was seeded at 5000 cells/well in sextuplicates in 96-well plates and incubated for 24h, 48h and 72h. Proliferation were measured using CyQuant Proliferation NF assay (Thermo Fisher Scientific), following manufacturers manual. Fluorescence was measured using Victor-3 multilabel reader (PerkinElmer), with excitation 485 nm and emission 530 nm. Three independent experiments were performed (SK-N-AS passage 22, 23 and 25, respectively).
Cell migration
Transfected HEK293 cells were plated in quadruplicates at 4x104 cells/well in Oris Cell Migration plates (Tebu-Bio) containing cell-seeding stopper. Cells were allowed to attach and reach 100% confluency during 48h and the cell stopper was removed. One well was photographed in a light microscope directly after removal of the cell stopper to get a starting area and all wells were photographed (5X) on Zeiss Axio Vert.A1 after 24h to determine the migration of the cells. The trial was repeated once, and only wells with cell layers showing an intact circular wound edge were included in the analysis (2–4 replicates/construct). Remaining area was measured with Image J v1.0 [74], and calculated as percent area left compared to well with starting area (100%).
Transferrin assay
Transfected SK-N-AS cells were plated (2.5x104) onto MilliCell EZ slides (Merck Millipore) and allowed to attach during 48 hours, then starved for 1,5 h in Life cell imaging solution (LCIS) (Thermo Fisher scientific). Cells were subsequently incubated for 15 minutes in 20 μg/ml Alexa fluor conjugated 555 human transferrin (T35352, Thermo Fisher) and thereafter fixed for 15 minutes in 4% paraformaldehyde solution (HistoLab). Chamber slide walls were detached and slides were mounted with Prolong Gold (Invitrogen, Life Technologies). Pictures were taken from 4–7 different areas of the slides with LSM700 confocal microscope (63X, Carl Zeiss) or Zeiss Axioscope 2 Plus fluorescence microscope equipped with Nikon DS- Qi1Mc camera (40X, Carl Zeiss, DAPI: 300 milliseconds (ms), Alexa fluor 555: 800 ms). For confocal settings see S1 File. Transferrin uptake was calculated in 4 areas by measuring total fluorescence staining from 40x pictures with Image J with Fiji package v1.0 [75], subtracted by the mean of background squares, and fluorescence was then divided by the number of cells in each picture.
Microarray expression analysis of SK-N-AS mutant clones
Stably transfected SK-N-AS cells were seeded onto T75 flasks (1x106 cells), then cultured and harvested at 24, 48 and 72 hours. The procedure was repeated twice (at passage 23 and passage 30). Cells were pelleted and RNA was extracted using Maxwell 16 LEV SimplyRNA kit (Promega) according to manufacturer’s protocol. RNA quality assessment was performed by measuring absorbance (A260/280; 1.9–2.1) on Nanodrop (Denovix DS-11 spectrophotometer) and RNA integrity number (RIN) values (>9.0) with High Sensitivity RNA Screen tape on 2200 Tape station (Agilent Technologies). Expression analysis was performed with Clariom S arrays for five constructs (empty vector, MYO5BWT, p.L587P, p.G1611S, and p.R1641C) at three time points of proliferation (24h, 48h, 72h) in duplicates by Eurofins Genomics (www.eurofinsgenomics.eu). Expression data on the 40 top-ranked differentially expressed genes from all three MYO5B mutants, i.e. genes present as differential expressed top-candidates in at least two out of three time points in at least two mutants, or at least in one time point in all three mutants, are presented in S5 Table. Next, Gene Ontology analysis by GOrilla (http://cbl-gorilla.cs.technion.ac.il [38]) and gene set enrichment analysis (GSEA, [39,40]) was performed on the gene lists. For further details on microarray and enrichment analyses see S1 File.
Verification by quantitative real-time PCR (qPCR)
From the top-ranked genes in the microarray experiment, the 29 most differentially expressed genes in all three mutants were selected; absolute mean log2 fold change > 0.58 (corresponding to a 1.5 relative fold change) when compared to MYO5BWT. cDNA was synthesized using High-Capacity RNA-to-cDNA Kit (ThermoFisher Scientific) with 200 ng of extracted total RNA from the five stably transfected SK-N-AS constructs from four experiments each (p21, p23, p28, p30) at proliferation time point of 48h. Using custom-designed TaqMan Array Cards, qPCR analysis was performed for 32 genes (29 genes of interest, and 3 endogenous controls) in triplicates, run on a QuantStudio 12K Flex Real-Time PCR System according to manufacturer’s instruction (ThermoFisher Scientific). The three endogenous controls; GAPDH, RPL37A, TBP, were selected based on their low variance between samples from the expression microarray analysis. The design of the TaqMan Array Micro Fluidic Card is presented in S8 Table. The differential expression between MYO5B mutants versus MYO5BWT constructs in each passage was calculated according to the ΔΔCt-method with normalization to the geometric mean of the three endogenous control genes.
Statistical analysis
The protein expression of MYO5B by IHC and mRNA expression by microarray for metastatic versus non-metastatic tumors was analyzed using Mann-Whitney and t-test respectively (Table 1). Proliferation data points were tested for normality using Kolmogorv-Smirnov test (p>0.05 in all SK-N-AS clones and all time points), and proliferation was analyzed with paired t-test including the mean from each trial at 24, 48 and 72 hours. Western blot of TfR in SK-N-AS was analyzed with ANOVA, followed by Fisher’s LSD comparing MYO5-mutants to MYO5BWT and empty vector. All statistical analyses were executed in IBM SPSS Statistics version 25.
Supporting information
Staining of Tyrosine hydroxylase (TH) in 23 patient tumor sections. Upper left panel shows a healthy adrenal gland (cortex and medulla) as a positive control. All tumor sections were positive for TH (21 cases), except for CPN3 and CPN12 which were positive for synaptophysin and chromogranin A. Pictures are scanned at 20X magnification using a Leica SCN400 scanner, scale bar shown is 50μm.
(TIF)
Two major sample clusters appear; cluster 1with pseudohypoxic signaling (SDHB, VHL, and EPAS1 tumors) and cluster 2 with kinase signaling (RET, NF1 and HRAS tumors). The heat map color scale is based on standard deviations (sd) and ranges from +2 sd (red) to -2 sd (green). The status of malignancy and diagnosis are shown by grey and black squares; black = metastatic, dark grey = multifocal, light grey = non-metastatic; black = PGL, grey = PCC. Mutation status is marked as follows: red = EPAS1, orange = SDHB, yellow = VHL, blue = RET, turquoise = NF1, green = HRAS, brown = SDHA, white = not determined (nd). Two cases in cluster 2, one harboring a SDHB-mutation (CPN14) and one with no mutation identified (CPN93) showed NF1-manifestation.
(TIF)
Expression of 21 out of 29 most differentially expressed genes (A) and expression of 9 MYO5B-associated RABs (B) from the functional study of three MYO5B mutants. Tumor samples are displayed according to cluster subgroups. Genes are sorted by hierarchical clustering. The heat map color scale is based on standard deviations (sd) and ranges from +2 sd (red) to -2 sd (green). The status of malignancy and diagnosis are shown by grey and black squares; black = metastatic, dark grey = multifocal, light grey = non-metastatic; black = PGL, grey = PCC. Mutation status is marked as follows: red = EPAS1, orange = SDHB, yellow = VHL, blue = RET, turquoise = NF1, green = HRAS, brown = SDHA, white = not determined (nd).
(TIF)
The 40 genes for calling of mutations in exomes were selected based on the following criteria; previously established or reported with germline and/or recurrent somatic mutations in PPGL (Buffet 2018 [9], Cascon 2015 [10], Dahia 2014 [7], Dwight 2017 [71], Fishbein 2017 [4], Papathomas 2014 [11], Remacha 2018 [12], Remacha 2019 [13], Yang 2015 [15], Yeh [77]), or recurrently occurring genes between lists of several PPGL studies (Castro-Vega 2015 [70], Flynn 2015 [72], Juhlin 2015 [73], Fishbein 2015 [18]), or recurrently mutated within our previous study (Wilzén 2016 [14]). Also, MYO5B-paralogs MYO5A and MYO5C were included. The 14 most commonly mutated genes in PPGL are marked with a star (*).
(PDF)
Mutation analysis by Exome sequencing and MLPA (see material and methods for details). Paired tumor tissue and normal samples (T & N) or single tumor samples (T) run by different library preparation kits (SureSelect v3 or v5 or Clinical research exome (CRE v2)). Variant filtering was performed by Alissa Interpret (Agilent Technologies) and somatic filtering in paired samples (T-N) was according to Wilzen et al., 2016 [14]. Major variant: pathogenic or likely pathogenic mutation in any of the 14 PPGL susceptibility genes and their allele frequency (AF) in normal and/or tumor sample. Present in Database: Variants previously reported in ClinVar (www.ncbi.nlm.nih.gov/clinvar/) or HGMD (portal.biobase-international.com) databases. Variants were defined as germline if occurring in the normal blood/tissue sample, and as somatic if only occurring in the tumor tissue sample. Other variants: secondary variants present in the 40-gene set occurring in AF>0.2, predicted to be damaging by at least 2 out of 3 functional prediction software (Polyhen, SIFT, and MutationTaster), and present <0.1% (germline) or 0% (somatic) in normal population databases. *Variants previously reported in COSMIC were included at lower AF. nd = not determined.
(PDF)
The 153 genes with highest variance in 26 tumors samples, discriminating two expression clusters of PPGL tumors.
(PDF)
Microarray expression analysis of SK-N-AS constructs; MUT 1(p.L587P), MUT 2 (p.G1611S), and MUT 3 (p.R1641C) and wild type (WT) MYO5B compared to empty vector (EV). Calculations are based on measures from two SK-N-AS passages (p23 and p30) for each mutation and time point of proliferation (24h, 48h and 72h). t-statistic (t), significance (P.Value, and adj.p) and fold change (FC) from MUTvsEV group comparison.
(PDF)
Average expression values (log2-transformed) from microarray mRNA differential expression analysis of three MYO5B mutants in SK-N-AS cells; MUT 1(p.L587P), MUT 2 (p.G1611S), and MUT 3 (p.R1641C) compared to wild type (WT) MYO5B. Calculations are based on measures from two trials and three time points of proliferation (SK-N-AS passages p23 and p30 for time point 48h and 72h, and one replicated SK-N-AS p23 trial at 24h). t-statistic (t), significance (P.Value, and adj.p) and fold change (FC) from the MUTvsWT group comparisons are presented per mutation and time point. Only the top -ranked genes, i.e. present in at least 2 time points for at least 2 mutants, or at least 1 time point for 3 mutants, are listed in the table (see S1 File for details).
(XLSX)
The top-ranked differentially expressed genes (see S5 Table) from all three MYO5B mutants were analyzed using GOrilla (Gene Ontology enRIchment anaLysis and visuaLizAtion tool, http://cbl-gorilla.cs.technion.ac.il) with all genes present on the microarray used as background. N = total no. of genes recognized by Gorilla and associated with GO terms, B = no. of genes associated with the specified GO term, n = no. of genes recognized by GOrilla in the target set (one gene annotation including several genes and one gene not associated with any GO terms were excluded), b = no. of genes in the target set associated with the specified GO term. P-value threshold < 10^-3.
(PDF)
Gene Set Enrichment Analysis (GSEA) performed on expression gene lists (ranked after mean fold change) for each mutant; MUT1 (p.L587P), MUT2 (p.G1611S), and MUT3 (p.R1641C), using 236 gene sets (50 H Hallmark, and 186 C2: KEGG curated gene sets) from the Molecular Signature database (http://software.broadinstitute.org/gsea/msigdb/index.jsp). The overlapping gene sets among the 20 top-ranked with up-regulated genes (UP) and 20 top-ranked with down-regulated genes (DOWN) for each mutations are listed.
(PDF)
Assay-ID and target information for TaqMan primers and probes used for qPCR verification of the 29 most differentially expressed genes in MYO5B mutants. INV = Inventoried assay, MTO = Made-to-order assay.
(PDF)
(PDF)
Acknowledgments
We would like to thank Gülay Altiparmak at Sahlgrenska Cancer Centre at University of Gothenburg for preparing histopathological material, Angela Martinez-Monleon/Tommy Martinsson at Clinical Genetics at the University of Gothenburg for the SK-N-AS cell line, the Center for Cellular Imaging at Sahlgrenska Academy, Gothenburg University for access and support using their confocal microscope. Gene Core Facility at Sahlgrenska Hospital for access to Illumina sequencing systems, and the Clinical Genomics Gothenburg unit at Sahlgrenska Academy, Gothenburg University for exome-sequencing data management and support.
Data Availability
Somatic sequence data (whole exome) has been deposited at the European Genome-phenome Archive (EGA, https://ega-archive.org), which is hosted by the EBI and the CRG, under accession number EGAS00001001601 and EGAS00001003991. For data access some restrictions will apply to fulfil ethical agreement and informed consent of patients. Germline data cannot be shared publicly due to patient confidentiality. For data access requests, contact the data access committee: dac-cgnt@gu.se.
Funding Statement
This work was supported by the Swedish Cancer Society (www.cancerfonden.se, grant numbers 2018/825 to FA and CAN 2017/742 to ONW), the Swedish Children’s Cancer Foundation (www.barncancerfonden.se, grant numbers PR2016-0016 and PR2017-0029 to FA and NC2014-0036 to TT), the Assar Gabrielsson Foundation (www.agfond.se, grant numbers FB 15–75 to TT), ALF-agreement (www.researchweb.org/is/alfgbg, ALFGBG-725101 to ONW). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wangberg B, Muth A, Khorram-Manesh A, Jansson S, Nilsson O, Forssell-Aronsson E, et al. Malignant pheochromocytoma in a population-based study: survival and clinical results. Ann N Y Acad Sci. 2006;1073:512–6. 10.1196/annals.1353.054 [DOI] [PubMed] [Google Scholar]
- 2.Kolackov K, Tupikowski K, Bednarek-Tupikowska G. Genetic aspects of pheochromocytoma. Adv Clin Exp Med. 2012;21(6):821–9. [PubMed] [Google Scholar]
- 3.Goldstein RE, O'Neill JA Jr., Holcomb GW 3rd, Morgan WM 3rd, Neblett WW 3rd, Oates JA, et al. Clinical experience over 48 years with pheochromocytoma. Ann Surg. 1999;229(6):755–64; discussion 64–6. 10.1097/00000658-199906000-00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fishbein L, Leshchiner I, Walter V, Danilova L, Robertson AG, Johnson AR, et al. Comprehensive Molecular Characterization of Pheochromocytoma and Paraganglioma. Cancer Cell. 2017;31(2):181–93. 10.1016/j.ccell.2017.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hescot S, Leboulleux S, Amar L, Vezzosi D, Borget I, Bournaud-Salinas C, et al. One-year progression-free survival of therapy-naive patients with malignant pheochromocytoma and paraganglioma. J Clin Endocrinol Metab. 2013;98(10):4006–12. 10.1210/jc.2013-1907 [DOI] [PubMed] [Google Scholar]
- 6.Favier J, Amar L, Gimenez-Roqueplo AP. Paraganglioma and phaeochromocytoma: from genetics to personalized medicine. Nat Rev Endocrinol. 2015;11(2):101–11. 10.1038/nrendo.2014.188 [DOI] [PubMed] [Google Scholar]
- 7.Dahia PL. Pheochromocytoma and paraganglioma pathogenesis: learning from genetic heterogeneity. Nat Rev Cancer. 2014;14(2):108–19. 10.1038/nrc3648 [DOI] [PubMed] [Google Scholar]
- 8.Muth A, Abel F, Jansson S, Nilsson O, Ahlman H, Wangberg B. Prevalence of germline mutations in patients with pheochromocytoma or abdominal paraganglioma and sporadic presentation: a population-based study in Western Sweden. World J Surg. 2012;36(6):1389–94. 10.1007/s00268-012-1430-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buffet A, Morin A, Castro-Vega LJ, Habarou F, Lussey-Lepoutre C, Letouze E, et al. Germline Mutations in the Mitochondrial 2-Oxoglutarate/Malate Carrier SLC25A11 Gene Confer a Predisposition to Metastatic Paragangliomas. Cancer Res. 2018;78(8):1914–22. 10.1158/0008-5472.CAN-17-2463 [DOI] [PubMed] [Google Scholar]
- 10.Cascon A, Comino-Mendez I, Curras-Freixes M, de Cubas AA, Contreras L, Richter S, et al. Whole-exome sequencing identifies MDH2 as a new familial paraganglioma gene. J Natl Cancer Inst. 2015;107(5). [DOI] [PubMed] [Google Scholar]
- 11.Papathomas TG, Oudijk L, Zwarthoff EC, Post E, Duijkers FA, van Noesel MM, et al. Telomerase reverse transcriptase promoter mutations in tumors originating from the adrenal gland and extra-adrenal paraganglia. Endocr Relat Cancer. 2014;21(4):653–61. 10.1530/ERC-13-0429 [DOI] [PubMed] [Google Scholar]
- 12.Remacha L, Curras-Freixes M, Torres-Ruiz R, Schiavi F, Torres-Perez R, Calsina B, et al. Gain-of-function mutations in DNMT3A in patients with paraganglioma. Genet Med. 2018;20(12):1644–51. 10.1038/s41436-018-0003-y [DOI] [PubMed] [Google Scholar]
- 13.Remacha L, Pirman D, Mahoney CE, Coloma J, Calsina B, Curras-Freixes M, et al. Recurrent Germline DLST Mutations in Individuals with Multiple Pheochromocytomas and Paragangliomas. Am J Hum Genet. 2019;104(5):1008–10. 10.1016/j.ajhg.2019.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilzen A, Rehammar A, Muth A, Nilsson O, Tesan Tomic T, Wangberg B, et al. Malignant pheochromocytomas/paragangliomas harbor mutations in transport and cell adhesion genes. Int J Cancer. 2016;138(9):2201–11. 10.1002/ijc.29957 [DOI] [PubMed] [Google Scholar]
- 15.Yang C, Zhuang Z, Fliedner SM, Shankavaram U, Sun MG, Bullova P, et al. Germ-line PHD1 and PHD2 mutations detected in patients with pheochromocytoma/paraganglioma-polycythemia. J Mol Med (Berl). 2015;93(1):93–104. 10.1007/s00109-014-1205-7 [DOI] [PubMed] [Google Scholar]
- 16.Jimenez C, Rohren E, Habra MA, Rich T, Jimenez P, Ayala-Ramirez M, et al. Current and future treatments for malignant pheochromocytoma and sympathetic paraganglioma. Curr Oncol Rep. 2013;15(4):356–71. 10.1007/s11912-013-0320-x [DOI] [PubMed] [Google Scholar]
- 17.Comino-Mendez I, Gracia-Aznarez FJ, Schiavi F, Landa I, Leandro-Garcia LJ, Leton R, et al. Exome sequencing identifies MAX mutations as a cause of hereditary pheochromocytoma. Nat Genet. 2011;43(7):663–7. 10.1038/ng.861 [DOI] [PubMed] [Google Scholar]
- 18.Fishbein L, Khare S, Wubbenhorst B, DeSloover D, D'Andrea K, Merrill S, et al. Whole-exome sequencing identifies somatic ATRX mutations in pheochromocytomas and paragangliomas. Nature communications. 2015;6:6140 10.1038/ncomms7140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson LD. Pheochromocytoma of the Adrenal gland Scaled Score (PASS) to separate benign from malignant neoplasms: a clinicopathologic and immunophenotypic study of 100 cases. Am J Surg Pathol. 2002;26(5):551–66. 10.1097/00000478-200205000-00002 [DOI] [PubMed] [Google Scholar]
- 20.Ouderkirk JL, Krendel M. Non-muscle myosins in tumor progression, cancer cell invasion, and metastasis. Cytoskeleton (Hoboken). 2014;71(8):447–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lapierre LA, Kumar R, Hales CM, Navarre J, Bhartur SG, Burnette JO, et al. Myosin vb is associated with plasma membrane recycling systems. Mol Biol Cell. 2001;12(6):1843–57. 10.1091/mbc.12.6.1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schafer JC, Baetz NW, Lapierre LA, McRae RE, Roland JT, Goldenring JR. Rab11-FIP2 interaction with MYO5B regulates movement of Rab11a-containing recycling vesicles. Traffic. 2014;15(3):292–308. 10.1111/tra.12146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roland JT, Bryant DM, Datta A, Itzen A, Mostov KE, Goldenring JR. Rab GTPase-Myo5B complexes control membrane recycling and epithelial polarization. Proc Natl Acad Sci U S A. 2011;108(7):2789–94. 10.1073/pnas.1010754108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kravtsov D, Mashukova A, Forteza R, Rodriguez MM, Ameen NA, Salas PJ. Myosin 5b loss of function leads to defects in polarized signaling: implication for microvillus inclusion disease pathogenesis and treatment. Am J Physiol Gastrointest Liver Physiol. 2014;307(10):G992–G1001. 10.1152/ajpgi.00180.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muller T, Hess MW, Schiefermeier N, Pfaller K, Ebner HL, Heinz-Erian P, et al. MYO5B mutations cause microvillus inclusion disease and disrupt epithelial cell polarity. Nat Genet. 2008;40(10):1163–5. 10.1038/ng.225 [DOI] [PubMed] [Google Scholar]
- 26.van der Velde KJ, Dhekne HS, Swertz MA, Sirigu S, Ropars V, Vinke PC, et al. An overview and online registry of microvillus inclusion disease patients and their MYO5B mutations. Hum Mutat. 2013;34(12):1597–605. 10.1002/humu.22440 [DOI] [PubMed] [Google Scholar]
- 27.Thoeni CE, Vogel GF, Tancevski I, Geley S, Lechner S, Pfaller K, et al. Microvillus inclusion disease: loss of Myosin vb disrupts intracellular traffic and cell polarity. Traffic. 2014;15(1):22–42. 10.1111/tra.12131 [DOI] [PubMed] [Google Scholar]
- 28.Li YR, Yang WX. Myosin superfamily: The multi-functional and irreplaceable factors in spermatogenesis and testicular tumors. Gene. 2016;576(1 Pt 2):195–207. [DOI] [PubMed] [Google Scholar]
- 29.Lan L, Han H, Zuo H, Chen Z, Du Y, Zhao W, et al. Upregulation of myosin Va by Snail is involved in cancer cell migration and metastasis. Int J Cancer. 2010;126(1):53–64. 10.1002/ijc.24641 [DOI] [PubMed] [Google Scholar]
- 30.Dong W, Chen X, Chen P, Yue D, Zhu L, Fan Q. Inactivation of MYO5B promotes invasion and motility in gastric cancer cells. Digestive diseases and sciences. 2012;57(5):1247–52. 10.1007/s10620-011-1989-z [DOI] [PubMed] [Google Scholar]
- 31.Dong W, Wang L, Shen R. MYO5B is epigenetically silenced and associated with MET signaling in human gastric cancer. Digestive diseases and sciences. 2013;58(7):2038–45. 10.1007/s10620-013-2600-6 [DOI] [PubMed] [Google Scholar]
- 32.Letellier E, Schmitz M, Ginolhac A, Rodriguez F, Ullmann P, Qureshi-Baig K, et al. Loss of Myosin Vb in colorectal cancer is a strong prognostic factor for disease recurrence. Br J Cancer. 2017;117(11):1689–701. 10.1038/bjc.2017.352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knowles BC, Roland JT, Krishnan M, Tyska MJ, Lapierre LA, Dickman PS, et al. Myosin Vb uncoupling from RAB8A and RAB11A elicits microvillus inclusion disease. J Clin Invest. 2014;124(7):2947–62. 10.1172/JCI71651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rief M, Rock RS, Mehta AD, Mooseker MS, Cheney RE, Spudich JA. Myosin-V stepping kinetics: a molecular model for processivity. Proc Natl Acad Sci U S A. 2000;97(17):9482–6. 10.1073/pnas.97.17.9482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klein M, Vignaud JM, Hennequin V, Toussaint B, Bresler L, Plenat F, et al. Increased expression of the vascular endothelial growth factor is a pejorative prognosis marker in papillary thyroid carcinoma. J Clin Endocrinol Metab. 2001;86(2):656–8. 10.1210/jcem.86.2.7226 [DOI] [PubMed] [Google Scholar]
- 36.Dahia PL, Ross KN, Wright ME, Hayashida CY, Santagata S, Barontini M, et al. A HIF1alpha regulatory loop links hypoxia and mitochondrial signals in pheochromocytomas. PLoS Genet. 2005;1(1):72–80. 10.1371/journal.pgen.0010008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evenepoel L, Papathomas TG, Krol N, Korpershoek E, de Krijger RR, Persu A, et al. Toward an improved definition of the genetic and tumor spectrum associated with SDH germ-line mutations. Genet Med. 2015;17(8):610–20. 10.1038/gim.2014.162 [DOI] [PubMed] [Google Scholar]
- 38.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48 10.1186/1471-2105-10-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varemo L, Nielsen J, Nookaew I. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res. 2013;41(8):4378–91. 10.1093/nar/gkt111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Astuti D, Morris M, Krona C, Abel F, Gentle D, Martinsson T, et al. Investigation of the role of SDHB inactivation in sporadic phaeochromocytoma and neuroblastoma. Br J Cancer. 2004;91(10):1835–41. 10.1038/sj.bjc.6602202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.El-Badry OM, Romanus JA, Helman LJ, Cooper MJ, Rechler MM, Israel MA. Autonomous growth of a human neuroblastoma cell line is mediated by insulin-like growth factor II. J Clin Invest. 1989;84(3):829–39. 10.1172/JCI114243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rapizzi E, Ercolino T, Fucci R, Zampetti B, Felici R, Guasti D, et al. Succinate dehydrogenase subunit B mutations modify human neuroblastoma cell metabolism and proliferation. Horm Cancer. 2014;5(3):174–84. 10.1007/s12672-014-0172-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rapizzi E, Fucci R, Giannoni E, Canu L, Richter S, Cirri P, et al. Role of microenvironment on neuroblastoma SK-N-AS SDHB-silenced cell metabolism and function. Endocr Relat Cancer. 2015;22(3):409–17. 10.1530/ERC-14-0479 [DOI] [PubMed] [Google Scholar]
- 45.Royer C, Lu X. Epithelial cell polarity: a major gatekeeper against cancer? Cell Death Differ. 2011;18(9):1470–7. 10.1038/cdd.2011.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Izidoro-Toledo TC, Borges AC, Araujo DD, Mazzi DP, Nascimento Junior FO, Sousa JF, et al. A myosin-Va tail fragment sequesters dynein light chains leading to apoptosis in melanoma cells. Cell Death Dis. 2013;4:e547 10.1038/cddis.2013.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li YR, Yang WX. Myosins as fundamental components during tumorigenesis: diverse and indispensable. Oncotarget. 2016;7(29):46785–812. 10.18632/oncotarget.8800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Daniels TR, Bernabeu E, Rodriguez JA, Patel S, Kozman M, Chiappetta DA, et al. The transferrin receptor and the targeted delivery of therapeutic agents against cancer. Biochim Biophys Acta. 2012;1820(3):291–317. 10.1016/j.bbagen.2011.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olsson M, Beck S, Kogner P, Martinsson T, Caren H. Genome-wide methylation profiling identifies novel methylated genes in neuroblastoma tumors. Epigenetics. 2016;11(1):74–84. 10.1080/15592294.2016.1138195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim J, Kim WH, Byeon SJ, Lee BL, Kim MA. Epigenetic Downregulation and Growth Inhibition of IGFBP7 in Gastric Cancer. Asian Pac J Cancer Prev. 2018;19(3):667–75. 10.22034/APJCP.2018.19.3.667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akiel M, Guo C, Li X, Rajasekaran D, Mendoza RG, Robertson CL, et al. IGFBP7 Deletion Promotes Hepatocellular Carcinoma. Cancer Res. 2017;77(15):4014–25. 10.1158/0008-5472.CAN-16-2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chuanyu S, Yuqing Z, Chong X, Guowei X, Xiaojun Z. Periostin promotes migration and invasion of renal cell carcinoma through the integrin/focal adhesion kinase/c-Jun N-terminal kinase pathway. Tumour Biol. 2017;39(4):1010428317694549 10.1177/1010428317694549 [DOI] [PubMed] [Google Scholar]
- 53.Gonzalez-Gonzalez L, Alonso J. Periostin: A Matricellular Protein With Multiple Functions in Cancer Development and Progression. Front Oncol. 2018;8:225 10.3389/fonc.2018.00225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cai T, Chen X, Wang R, Xu H, You Y, Zhang T, et al. Expression of insulinoma-associated 2 (INSM2) in pancreatic islet cells is regulated by the transcription factors Ngn3 and NeuroD1. Endocrinology. 2011;152(5):1961–9. 10.1210/en.2010-1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang L, Sun ZS, Xiang B, Wei CJ, Wang Y, Sun K, et al. Targeted deletion of Insm2 in mice result in reduced insulin secretion and glucose intolerance. J Transl Med. 2018;16(1):297 10.1186/s12967-018-1665-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jyotsna VP, Malik E, Birla S, Sharma A. Novel MEN 1 gene findings in rare sporadic insulinoma—a case control study. BMC Endocr Disord. 2015;15:44 10.1186/s12902-015-0041-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yagi T, Kubota E, Koyama H, Tanaka T, Kataoka H, Imaeda K, et al. Glucagon promotes colon cancer cell growth via regulating AMPK and MAPK pathways. Oncotarget. 2018;9(12):10650–64. 10.18632/oncotarget.24367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sosnicki S, Kapral M, Weglarz L. Molecular targets of metformin antitumor action. Pharmacol Rep. 2016;68(5):918–25. 10.1016/j.pharep.2016.04.021 [DOI] [PubMed] [Google Scholar]
- 59.Daugan M, Dufay Wojcicki A, d'Hayer B, Boudy V. Metformin: An anti-diabetic drug to fight cancer. Pharmacol Res. 2016;113(Pt A):675–85. 10.1016/j.phrs.2016.10.006 [DOI] [PubMed] [Google Scholar]
- 60.Li M, Jiang X, Su T, Jiang L, Zhou W, Wang W. Metformin Suppresses Proliferation and Viability of Rat Pheochromocytoma Cells. Med Sci Monit. 2017;23:3253–60. 10.12659/msm.903348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lopez-Jimenez E, Gomez-Lopez G, Leandro-Garcia LJ, Munoz I, Schiavi F, Montero-Conde C, et al. Research resource: Transcriptional profiling reveals different pseudohypoxic signatures in SDHB and VHL-related pheochromocytomas. Mol Endocrinol. 2010;24(12):2382–91. 10.1210/me.2010-0256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jochmanova I, Pacak K. Pheochromocytoma: The First Metabolic Endocrine Cancer. Clin Cancer Res. 2016;22(20):5001–11. 10.1158/1078-0432.CCR-16-0606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Favier J, Briere JJ, Burnichon N, Riviere J, Vescovo L, Benit P, et al. The Warburg effect is genetically determined in inherited pheochromocytomas. PLoS One. 2009;4(9):e7094 10.1371/journal.pone.0007094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Berkel A, Rao JU, Kusters B, Demir T, Visser E, Mensenkamp AR, et al. Correlation between in vivo 18F-FDG PET and immunohistochemical markers of glucose uptake and metabolism in pheochromocytoma and paraganglioma. J Nucl Med. 2014;55(8):1253–9. 10.2967/jnumed.114.137034 [DOI] [PubMed] [Google Scholar]
- 65.van Lennep JR, Romijn JA, Harinck HI. Multi-organ failure after a glucagon test. Lancet. 2007;369(9563):798 10.1016/S0140-6736(07)60365-1 [DOI] [PubMed] [Google Scholar]
- 66.Hosseinnezhad A, Black RM, Aeddula NR, Adhikari D, Trivedi N. Glucagon-induced pheochromocytoma crisis. Endocr Pract. 2011;17(3):e51–4. 10.4158/EP10388.CR [DOI] [PubMed] [Google Scholar]
- 67.Legler A, Kim RK, Chawla N. Glucagon-induced hypertensive emergency: a case report. J Clin Anesth. 2016;35:493–6. 10.1016/j.jclinane.2016.08.033 [DOI] [PubMed] [Google Scholar]
- 68.Lenders JW, Pacak K, Huynh TT, Sharabi Y, Mannelli M, Bratslavsky G, et al. Low sensitivity of glucagon provocative testing for diagnosis of pheochromocytoma. J Clin Endocrinol Metab. 2010;95(1):238–45. 10.1210/jc.2009-1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lussey-Lepoutre C, Hollinshead KE, Ludwig C, Menara M, Morin A, Castro-Vega LJ, et al. Loss of succinate dehydrogenase activity results in dependency on pyruvate carboxylation for cellular anabolism. Nature communications. 2015;6:8784 10.1038/ncomms9784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Castro-Vega LJ, Letouze E, Burnichon N, Buffet A, Disderot PH, Khalifa E, et al. Multi-omics analysis defines core genomic alterations in pheochromocytomas and paragangliomas. Nature communications. 2015;6:6044 10.1038/ncomms7044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dwight T, Na U, Kim E, Zhu Y, Richardson AL, Robinson BG, et al. Analysis of SDHAF3 in familial and sporadic pheochromocytoma and paraganglioma. BMC Cancer. 2017;17(1):497 10.1186/s12885-017-3486-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Flynn A, Benn D, Clifton-Bligh R, Robinson B, Trainer AH, James P, et al. The genomic landscape of phaeochromocytoma. J Pathol. 2015;236(1):78–89. 10.1002/path.4503 [DOI] [PubMed] [Google Scholar]
- 73.Juhlin CC, Stenman A, Haglund F, Clark VE, Brown TC, Baranoski J, et al. Whole-exome sequencing defines the mutational landscape of pheochromocytoma and identifies KMT2D as a recurrently mutated gene. Genes Chromosomes Cancer. 2015;54(9):542–54. 10.1002/gcc.22267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–5. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–82. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Qiu YL, Gong JY, Feng JY, Wang RX, Han J, Liu T, et al. Defects in myosin VB are associated with a spectrum of previously undiagnosed low gamma-glutamyltransferase cholestasis. Hepatology. 2017;65(5):1655–69. 10.1002/hep.29020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yeh IT, Lenci RE, Qin Y, Buddavarapu K, Ligon AH, Leteurtre E, et al. A germline mutation of the KIF1B beta gene on 1p36 in a family with neural and nonneural tumors. Hum Genet. 2008;124(3):279–85. 10.1007/s00439-008-0553-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Staining of Tyrosine hydroxylase (TH) in 23 patient tumor sections. Upper left panel shows a healthy adrenal gland (cortex and medulla) as a positive control. All tumor sections were positive for TH (21 cases), except for CPN3 and CPN12 which were positive for synaptophysin and chromogranin A. Pictures are scanned at 20X magnification using a Leica SCN400 scanner, scale bar shown is 50μm.
(TIF)
Two major sample clusters appear; cluster 1with pseudohypoxic signaling (SDHB, VHL, and EPAS1 tumors) and cluster 2 with kinase signaling (RET, NF1 and HRAS tumors). The heat map color scale is based on standard deviations (sd) and ranges from +2 sd (red) to -2 sd (green). The status of malignancy and diagnosis are shown by grey and black squares; black = metastatic, dark grey = multifocal, light grey = non-metastatic; black = PGL, grey = PCC. Mutation status is marked as follows: red = EPAS1, orange = SDHB, yellow = VHL, blue = RET, turquoise = NF1, green = HRAS, brown = SDHA, white = not determined (nd). Two cases in cluster 2, one harboring a SDHB-mutation (CPN14) and one with no mutation identified (CPN93) showed NF1-manifestation.
(TIF)
Expression of 21 out of 29 most differentially expressed genes (A) and expression of 9 MYO5B-associated RABs (B) from the functional study of three MYO5B mutants. Tumor samples are displayed according to cluster subgroups. Genes are sorted by hierarchical clustering. The heat map color scale is based on standard deviations (sd) and ranges from +2 sd (red) to -2 sd (green). The status of malignancy and diagnosis are shown by grey and black squares; black = metastatic, dark grey = multifocal, light grey = non-metastatic; black = PGL, grey = PCC. Mutation status is marked as follows: red = EPAS1, orange = SDHB, yellow = VHL, blue = RET, turquoise = NF1, green = HRAS, brown = SDHA, white = not determined (nd).
(TIF)
The 40 genes for calling of mutations in exomes were selected based on the following criteria; previously established or reported with germline and/or recurrent somatic mutations in PPGL (Buffet 2018 [9], Cascon 2015 [10], Dahia 2014 [7], Dwight 2017 [71], Fishbein 2017 [4], Papathomas 2014 [11], Remacha 2018 [12], Remacha 2019 [13], Yang 2015 [15], Yeh [77]), or recurrently occurring genes between lists of several PPGL studies (Castro-Vega 2015 [70], Flynn 2015 [72], Juhlin 2015 [73], Fishbein 2015 [18]), or recurrently mutated within our previous study (Wilzén 2016 [14]). Also, MYO5B-paralogs MYO5A and MYO5C were included. The 14 most commonly mutated genes in PPGL are marked with a star (*).
(PDF)
Mutation analysis by Exome sequencing and MLPA (see material and methods for details). Paired tumor tissue and normal samples (T & N) or single tumor samples (T) run by different library preparation kits (SureSelect v3 or v5 or Clinical research exome (CRE v2)). Variant filtering was performed by Alissa Interpret (Agilent Technologies) and somatic filtering in paired samples (T-N) was according to Wilzen et al., 2016 [14]. Major variant: pathogenic or likely pathogenic mutation in any of the 14 PPGL susceptibility genes and their allele frequency (AF) in normal and/or tumor sample. Present in Database: Variants previously reported in ClinVar (www.ncbi.nlm.nih.gov/clinvar/) or HGMD (portal.biobase-international.com) databases. Variants were defined as germline if occurring in the normal blood/tissue sample, and as somatic if only occurring in the tumor tissue sample. Other variants: secondary variants present in the 40-gene set occurring in AF>0.2, predicted to be damaging by at least 2 out of 3 functional prediction software (Polyhen, SIFT, and MutationTaster), and present <0.1% (germline) or 0% (somatic) in normal population databases. *Variants previously reported in COSMIC were included at lower AF. nd = not determined.
(PDF)
The 153 genes with highest variance in 26 tumors samples, discriminating two expression clusters of PPGL tumors.
(PDF)
Microarray expression analysis of SK-N-AS constructs; MUT 1(p.L587P), MUT 2 (p.G1611S), and MUT 3 (p.R1641C) and wild type (WT) MYO5B compared to empty vector (EV). Calculations are based on measures from two SK-N-AS passages (p23 and p30) for each mutation and time point of proliferation (24h, 48h and 72h). t-statistic (t), significance (P.Value, and adj.p) and fold change (FC) from MUTvsEV group comparison.
(PDF)
Average expression values (log2-transformed) from microarray mRNA differential expression analysis of three MYO5B mutants in SK-N-AS cells; MUT 1(p.L587P), MUT 2 (p.G1611S), and MUT 3 (p.R1641C) compared to wild type (WT) MYO5B. Calculations are based on measures from two trials and three time points of proliferation (SK-N-AS passages p23 and p30 for time point 48h and 72h, and one replicated SK-N-AS p23 trial at 24h). t-statistic (t), significance (P.Value, and adj.p) and fold change (FC) from the MUTvsWT group comparisons are presented per mutation and time point. Only the top -ranked genes, i.e. present in at least 2 time points for at least 2 mutants, or at least 1 time point for 3 mutants, are listed in the table (see S1 File for details).
(XLSX)
The top-ranked differentially expressed genes (see S5 Table) from all three MYO5B mutants were analyzed using GOrilla (Gene Ontology enRIchment anaLysis and visuaLizAtion tool, http://cbl-gorilla.cs.technion.ac.il) with all genes present on the microarray used as background. N = total no. of genes recognized by Gorilla and associated with GO terms, B = no. of genes associated with the specified GO term, n = no. of genes recognized by GOrilla in the target set (one gene annotation including several genes and one gene not associated with any GO terms were excluded), b = no. of genes in the target set associated with the specified GO term. P-value threshold < 10^-3.
(PDF)
Gene Set Enrichment Analysis (GSEA) performed on expression gene lists (ranked after mean fold change) for each mutant; MUT1 (p.L587P), MUT2 (p.G1611S), and MUT3 (p.R1641C), using 236 gene sets (50 H Hallmark, and 186 C2: KEGG curated gene sets) from the Molecular Signature database (http://software.broadinstitute.org/gsea/msigdb/index.jsp). The overlapping gene sets among the 20 top-ranked with up-regulated genes (UP) and 20 top-ranked with down-regulated genes (DOWN) for each mutations are listed.
(PDF)
Assay-ID and target information for TaqMan primers and probes used for qPCR verification of the 29 most differentially expressed genes in MYO5B mutants. INV = Inventoried assay, MTO = Made-to-order assay.
(PDF)
(PDF)
Data Availability Statement
Somatic sequence data (whole exome) has been deposited at the European Genome-phenome Archive (EGA, https://ega-archive.org), which is hosted by the EBI and the CRG, under accession number EGAS00001001601 and EGAS00001003991. For data access some restrictions will apply to fulfil ethical agreement and informed consent of patients. Germline data cannot be shared publicly due to patient confidentiality. For data access requests, contact the data access committee: dac-cgnt@gu.se.